|
MOLECULAR
GENETICS... DNA is molecular basis
of inheritance
DNA
toons... |
DNA molecules are transcribed as "software" and blindly
copied by polymerase enzymes, as the "hardware"
for the next generation. If the copying of the
software by the hardware is a process that
allows for errors,
then the replication of the software by the
hardware is a process that is evolvable.
|
.....
Genes
---> Enzymes
---> Metabolism
(phenotype) |
|
Overview of the
Central Dogma (3 min)* |
|
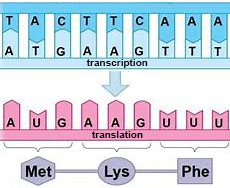
DNA --transcription--> RNA
--translation-->
Protein |
|
Central Dogma
of Molecular Biology* |
 |
the Era of OMIC's of modern
Molecular Biology - ["OME"
has the sense of
the whole]
Genomics
- study of the
genomes of organisms (sequencing, mapping,
& gene interactions)
Proteomics
- study of the
entire complement of proteins in a system or
organism
Transcriptomics
- all messenger RNA (mRNA) or
"transcripts," made in one cell or a
population of cells
Metabolomics - complete map of all
small-molecule metabolites in the human body
Structural Genomics -
determination of 10, 20, & 3D structure of all
proteins encoded by genome
BioInformatics
-
application of information technology to
field of molecular genetics- BIG DATA.
Pharmacogenomics
- genetic basis for
heritable & inter-individual variation in
drug responses
|

What is a GENE = ? How do we define
what a gene is???
- we seen that it is a basic unit of
information in all living
organisms
responsible for a morphological trait
(free ear lobes)...
- and DNA
is the likely chemical for this genetic
material, thus a gene is...
- a segment of DNA specifies a
morphological trait (round/wrinkled) |
- it's a discrete piece of deoxyribonucleic acid
that directs
the synthesis of a specific protein
- it's a linear
polymer of repeating nucleotide
monomers
nucleotides -->
A
adenine, C
cytosine
T
thymidine, G
guanine
yields -->
a polynucleotide
strand*
- structure
of polynucleotide strands of DNA &
RNA**view@home
What about,
retroviruses, as
HIV, Covid-19,
& TMV, which contain RNA,
that can molecularly
replicate, transcribe, and translate
RNA.
... maybe our definition of a gene
may need to be broader. |
 |

|
DNA and INFORMATIONAL
CONTENT PROCESSING
-
the letters of the genetic alphabet...
are just the 4
nucleotides A, T, G, &
C of DNA.
- the unit of genetic
information... or the
genetic 'word' is
called a CODON,
which is
a triplet
sequence of nucleotides, such as
p5'-CAT-3'p
in DNA makes a codon
&
these 3 nucleotides = 1 codon (word) & code for
an amino acid
in a polypeptide.
- thus the definition
of codon (a genetic
word) is an amino acid
5'-AUG-3' = 'MET'
(DNA) p5'-CAT-3'p
--> (mRNA)
5'-AUG-3' -->
(amino acid) --> N-methionine-C*
- Informational size of
a Human Genome:
≈ 3,088,286,401 base pairs [A:T & G:C] or
6.18 billion nucleotides in a human cell.
≈ 1,029,428,800 possible codons (amino acids "words")
in one human DNA strand.
average
page your textbook ≈
approx 850 words
thus, human genome is equal to 1,211,093
pages or
841 copies
of bio book (1440 pg)
reading at 3 bases/sec it would take you about 32.64 years @ a 24/7 rate
but, only about 80% or all DNA has any metabolic
function = 986,874 pages,
and only 1.5%
codes for proteins = 12,111 pages or 8.4 bio
books.
WOW... nanotechnology
animation*
& how
DNA is packaged*
► Of MICE and MEN... nearly every human
gene is also found in mice, and
98.8% of Chimp & Human DNA
is the same. A 1.2% difference is 35 million bp.
Many mammals including dogs, cats, rabbits, monkeys,
& apes have roughly
the same number of nucleotides in their genomes --
about 3 billion bp.
Estimates
suggest that Humans share 99.9% of their DNA person to person.

Experimental
Proof that DNA is the Genetic Material...
1. Bacterial Transformation
Experiments of
Fred Griffith
[a pneumonia vaccine
(1928)]...
he observed process whereby the bacterium
distinctly changed its form & function...
he
studied Streptococcus
pneumoniae -
pathogenic
S
strain & benign
R*
and showed the
transformation of R to S cells, via a genetic
transfer process,
he
called it a 'Transforming Principle'*,
converting one strain to a different strain.
2. Oswald Avery, Colin
MacLeod, & Maclyn
McCarty...
(pic - 1940's
Rockerfeller Institute, NY)
experimantally suggest the
transforming substance* is DNA molecules, but...
3.
Alfred
Hershey
& Martha Chase's (1952 Cold Spring Harbor, L.I.) virus replications
experiment...
Using a genetically controlled
biological activity, bacteriophage experiments*,
their inquiry based
experiments established DNA as the genetic basis
of heredity.
An
explanation of Hershey & Chase's
experiment = anim bacteriophage
exp*
the
use of radioisotopes in
biology* (32P)
DNA not (35S)
protein
guides viral replication
Conclusion
- DNA is
accepted as the Genetic Material...

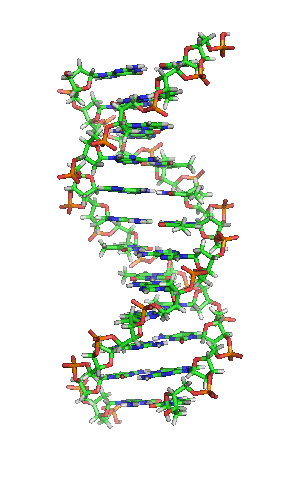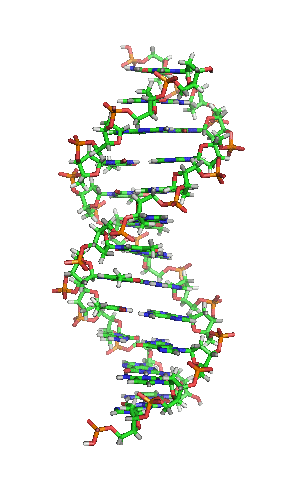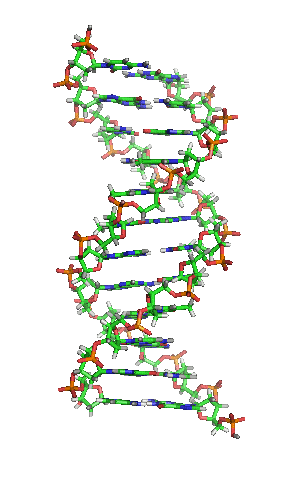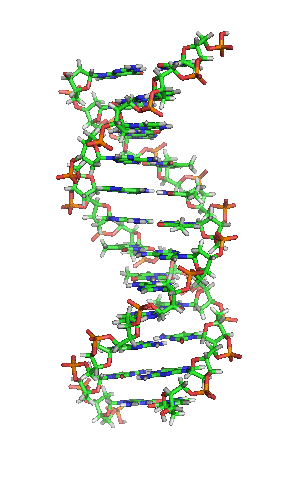
Structure of DNA...
Discovery of
Double Helix*
Watson's book
1962 Nobel prize
- JD Watson, Francis Crick, Maurice
Wilkins,
[??? but, what role did Erwin Chargaff & Rosy Franklin play???]
Dramatizations of DNA's
discovery: Race for the Double
Helix (1987) & Photograph
51 (2015)
and maybe one of the most Egregious Ripoff in the
History of
Science [by Howard Markel Oct
2021]
Watson & Crick, as
theoretical biologists, used two approaches to
decipher structure:
1. model building - figure* (are the bases
in/out*?)
2.
x-ray
diffraction*
and photo 51
favor a DNA helix of constant diameter*
we now know : DNA is a double
stranded*,
the helix*
of
polynucleotide chains,
made of 4 nucleotides - A,
T, G, C
(purine &
pyrimidines)
in 2 polynucleotide strands (polymer chains)
head-tail polarity [5'-----3'] - strands run antiparallel
held together via weak H-Bonds & complimentary pairing
-
Chargaff's
rule.... A:T**
G:C
A + G
/ T + C = 1.0
Fig's:
sugar-P-backbone
& base pairing*, (question*?),
dimensions*,
Summary animation of DNA Structure*view
 myDNAi timeline &
literature references --> [Nature:
171, 737-738(25 April 1953] myDNAi timeline &
literature references --> [Nature:
171, 737-738(25 April 1953]
view@home
REPLICATION OF DNA... copying of DNA into DNA
seems structurally obvious*??? figure
1. Patterns of
Replication* - 3 possibilities:
Conservative,
Semi-conservative,
& Dispersive
what was needed was a
procedure to tell difference between parental &
new DNA
Matt Meselson
& Frank Stahl 1958
- devised an experiment to establish
semi-conservative
replication of DNA - Explanations
of the Meslson-Stahl Experiment*view@home
the
experimental
design* to
separate parental
15N-DNA from 14N-new DNA-
(OLD vs. NEW DNA)?
the beauty of
their experiment is one
can predict what results ought to be before
doing it...
the results of the DNA centrifugation
experiment --> actual results* &
another fig-2
DNA Replication is
a semi-conservative
process.
view@home-1
2. DNA
POLYMERASE: [EC 2.7.7.7] copies DNA...
prokaryotic Pol I-IV eukaryotic α,β,γ,
& δ
bacterial polymerase is Pol III (pic) req: 4-deoxy-NTP's & a 3'-ssDNA template strand
DNA polymerase III reads template strand and
adds a complimentary nucleotide*view@home
reads 3' to 5' and synthesizes in 5' to 3'
direction --> templated polymerization*...
DNA
polymerase III proofreads* & the error rate of base
copying*
origin
of replication & bidirectional synthesis of
DNA* &
EM
pic*
the two DNA
strands are antiparallel (run opposite
directions) --> --> Overview
animation*3.5min
both strands are copied at
same time*
leading*
& lagging*
strand synthesis.
 |
|
|
|
the Model for studying the mechanisms of
DNA Replication is bacterial DNA
polymerase III...
all of the Enzymes* involved form a Replication
Complex called a Replisome:
the bacterial enzymes include*:
helicase
- untwists DNA
topoisomerase [DNA
gyrase] - removes
supercoils..... [animation]
single strand binding
proteins - stabilize replication fork,
Primase - makes RNA
primer terminus on leading strand & pieces on
lagging strand
POL III - synthesizes
new DNA strands
DNA polymerase I -
an exonuclease removes RNA primer, 1 base at a
time, & adds DNA bases
DNA ligase repairs Okazaki fragments (seals
lagging strand 3' open holes)
Structure
of DNA polymerase III Replisome*
copies both strands
simultaneously, as DNA is Threaded
Through a Replisome...
a "replication
machine", which may be stationary by
anchoring in nuclear matrix
Continuous & Discontinuous replication
occur simultaneously in both strands
Rates of DNA synthesis: myDNAi movie of-replication*view

END OF MATERIAL FOR
EXAM #4
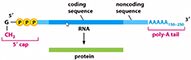
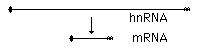
GENE Expression... DNA
--> RNA --> PROTEIN
the Central Dogma of Molecular
Biology describes
flow of genetic information*
DNA sequence
-------> RNA
sequence -----> amino acid sequence
CAT
AUG
MET
a triplet sequence in
DNA --> codon in mRNA -----> amino acid in protein
Information : triplet sequence in DNA
is the genetic word [a codon]
Transcription - copying of DNA
sequences into RNA
Translation - copying of RNA
sequences into protein
One Gene One Enzyme* - 1941
hypothesis that each gene directly
makes on enzyme (Beadle
& Tatum 1958 Nobel)
Compare Events:
Procaryotes* vs. Eucaryotes* = Separation of
labor
Differences DNA vs.
RNA (bases & sugars) and its single stranded

Transcription
is via RNA
polymerase...
RNA polymerase* transcribes
DNA
into an RNA product*.
- in bacteria
Sigma factor* binds promoter & initiates copying
of DNA*
- makes a
complimentary
copy* of just ONE*
of the two DNA strands
- activators &
transcription factors* are needed to recognize
specific DNA sequences
- Transcription factors* via protein motifs bind to
promoter DNA region
Transcription: the
Movie (short)*
and
Transcription: the
movie (long)*view@home
Cells make many KINDS of RNA
[table*]
tRNA -
small, 80n, anticodon sequence, single strand with 2ndary structure*
function = picks up aa & transports it to ribosome
[Mahlon
Hoagland]
rRNA - 3
individual pieces of RNA - make up the organelle = RIBOSOME
a
primary transcript of rRNA is processed
into the 3 pieces of
recall structure of
ribosome &
rRNApieces*

Other Classes of RNA Molecules
include:
snRNP's small nuclear RNA - plays a structural and catalytic
role in
spliceosome*
called "snurps"... there are 5 making up a
spliceosome [U1,
U2, U4, U5, & U6],
all urideine
rich - they participate in several RNA-RNA and
RNA-protein interactions
SRP (Signal
Recognition Particle):
contains a small cytoplasmic
7s-scRNA
component as part of the receptor complex,
which recognizes the signal
sequence
of polypeptides targeted to the ER -
figure*
snoRNA (small
nucleolar RNA) - are noncoding RNAs
located in the nucleolus, which
are involved in rRNA modifications by chemically
modifying them via methylation
and pseudouridylations
to generate mature RNA molecules...
Micro
RNAs (& siRNA): function
in gene
silencing & gene regulation.
are
single-stranded RNA molecules of 20-24
nucleotides in length, that are not
translated
into
protein (non-coding RNA) and form short stem-loop structures that
are partially
complimentary
to mRNAs & that
can block
translation* by
downregulating gene expressions.
FDA approves 1st
RNAi drug treatment
(august 2018) for ONPATTRO
a therapeutic that
treats
polyneuropathy in hereditary mutation in
amyloidogenic transthyretin amyloidosis
which
results in protein misfolds that form aggregates
and deposits amyloid plaque
in
multiple tissue and organs in the body.

TRANSLATION - Protein Synthesis... basics of translation
animation*
process of making a
protein in a specific amino
acid sequence
from a unique
mRNA sequence... [ Model*
& process* ]
polypeptides are built on the ribosome on a polysome.
SEQUENCE DETAILS...
[the Discovery of tRNA by Paul
Zamecnik - JBC article 1958]
1st
add an amino acid to tRNA -- > tRNA* --> aa-tRNA -
ACTIVATION + animation*
2nd assemble players [ribosome, mRNA, aa-tRNA]
- INITIATION
+ animation*
3rd adding new aa's via
peptidyl transferase
- ELONGATION*
+ animation
*
4th
stopping the process
-
TERMINATION + animation*
Review the processes: RNA Processing summary &
translation
summary fig*
initiation, elongation, & termination in animations
Real-time
animation of translation*view@home
| GENETIC CODE... is the genetic
alphabet spelling the genetic information
into proteins. |
..is the
sequence of nucleotides in DNA, but because mRNA
sequences specify sequence of amino acids in
a protein the code is
routinely shown as a mRNA CODE
|
 |
Coding
ratio
- how many nucleotides are
needed to specify 1 amino
acid ?
1n = 4
singlets, 2n
= 16 doublets, 3n = 64 triplet
codons a Triplet Code*
S.
Ochoa (NYU) (1959 Nobel) -
discovered polynucleotide
phosphorylase (it's an exonuclease)
Np-Np-Np-Np ◄----► Np-Np-Np + Np
but,
can be reveresed to make SYNTHETIC
mRNA
Marshall Nirenberg
(1968 Nobel) - made synthetic
mRNA's using
in vitro test tube systems
5'-UUU-3' = phe U
+ C --> UUU, UUC, UCC,
CCC
UCU, CUC, CCU, CUU =
mixes gave codons
the Genetic CODE* -
has 64 possible mRNA
triplet codons: new view of genetic code*
61 code for an amino
acid, 1
is an initiator (start)
codon
(AUG = nf-MET),
and
3 are molecular 'periods' or stop codons (UGA, UAA, & UAG);
Code is universal, redundant, but non-ambiguous, and exhibits "wobble*".
 "the sequencing of DNA
of dozens of species, from viruses to humans,
documents that "the sequencing of DNA
of dozens of species, from viruses to humans,
documents that
we're
all connected to the commonality of the
genetic code in evolution" jcv
(2007)
GENETIC CHANGE... a
change in DNA
nucleotide sequence
may equal a change
in mRNA)
- done in 2 significant natural ways...
via mutation
& recombination
[glossary]◄
1. MUTATION - a permanent change in a
cell's and progeny's DNA*that
results in
a different codon
may equal different amino
acid sequence
in a protein
Point mutation* - a single to
few nucleotides change...
- rate: E. coli = 1/105
per generation and humans = 1 in every
1010 base pairs.
ex: - deletions, insertions, frame-shift mutations [CAT]
= proof of a triplet codon
-
trinucleotide repeats - CAG repeats in
Huntington's disease...
single nucleotide base
substitutions* :
non-sense = change to
no amino acid [STOP
codon] UCA -->
UAA ser to non
mis-sense = different
amino
acid
UCA -->
UUA ser to leu
Sickle
Cell Anemia* - a mis-sense
mutation, where
one base pair
change = multiple effects in SCA-pleiotropy*
other point mutation
examples:
1) in
a blood disease, thalassemia,
a form of anemia
(change
at intron site)
2) mutant
AMPD1 which can lead to muscle cramps &
early fatigue
adenosine-monophosphate-deaminase-1 = is a
C to a T change
EFFECT
= no effect, detrimental
(lethal), +/-
functionality, beneficial

2. Recombinant DNA...
DNA made artificially
by combining DNA's from different cells
using laboratory techniques that often insert
NEW (foreign) DNA into recipient cells.
Natural
Recomination processes:
1. Fertilization* - sperm inserted into recipient
egg cell -->
zygote [n +
n = 2n]
2. exchange of
homologous chromatids via Crossing
over*
= new gene combo's
3. Transformation*
-
absorption of 'foreign' DNA by recipient
cells changes a cell
4. BACTERIAL
CONJUGATION* - involves
DNA
plasmids* (F+ or R = resistance)
Conjugation
may
result in genetic recombination in
bacteria [Hfr-CONJUGATION*]
5. virus Replication*
has
2 phases = lysis
vs. lysogeny*
&
[figure]
TRANSDUCTION* is transfer of foreign DNA into a cell via a viral vector
general transduction -
pieces of bacterial DNA are
packaged
w viral DNA during viral replication & lead
to recombination
restricted transduction
- a temperate phage goes lytic
carrying adjacent bacterial DNA into virus particle
& recombination
bacteriophages shuttle
bacterial genes between ecosystems

6. Biotechnology - Genetic Engineering - Designer Genes
RECOMBINANT
DNA TECHNOLOGY... involves
laboratory-made DNA molecules
and a
collection of experimental techniques,
which allow for the isolation, copying,
and
insertion of new DNA sequences into
host-recipient cells by a number of
unique
laboratory
protocols &
methodologies.
Restriction Endonucleases - glossary enzymes Cut DNA
at unique sequences*
...
Group of bacterial enzyme that cut DNA with 2
incisions through sugar-P's making small
pieces about 15-20,000 np's each; cuts are unequal or
blunt*
@ unique DNA sequences.
... To date some 3,000 restriction endonucleases
are known that recognize 250 cut sites
and some 800 restriction endonuclease are
available commercially.
-
Werner
Arber, Daniel Nathans, & Hamilton O. Smith
1978 Nobel
Eco-R1* - figure*
@ many are palindromes... ["never
odd or even"]
cartoon
cartoon
↔
5' G.AATTC
3'
5' G . . . . .
+ AATTC
3'
3' CTTAA.G
5'
3' CTTAA
. . . . G 5'
DNA's cut this way have STICKY
(complimentary)
ENDS &
can be spliced*
with other DNA molecules to make Recombinant DNA*
producing
new genes combos
and manufacture proteins for
medical use.
 Anecdote: Origins of Recombinant DNA Technology*
Anecdote: Origins of Recombinant DNA Technology*
Some Typical
Procedures in Biotechnology
(Genetic
Engineering)...
involving
restriction enzymes &
A:T G:C pairings
A.
Techniques involved in Cloning
a Gene...
1. via a plasmid* [ human shotgun plasmid
cloning*
&
animationview@home]
2. Librariesg...
[ whole genome library*
& animationview@home]
3.
cDNA genes*
[ synthetic genes made from mRNA
& cDNA
library* ]
4.
Probesg...
[ a
DNA probe of interest* + synthetic
DNA & Hybridization &
DNA Probes*
(how to find a gene
w/a probe + cDNAg
+ reverse transcriptaseg)
5. Polymerase Chain Raction
CR anim*
is a
method to amplify
small pieces of DNA
Taq
pol* & PCR
image
&
Covid PCR test*
& a
PCR long animview@home
Bio-Rad 1000 Thermal Cycler
& the PCR Song
& a cartoon
& "We are
the World"

B. Visualization of
DNA... to
locate a gene (or its activity) -->
Restriction Maps.
and other
practical applications in Forensics & Gene
Expression.
1. Restriction
Mapsg... the banding pattern seen in
gel electropherograms by
DNA treated with restriction enzymes and then
electrophoresed = "a
fingerprint"
electrophoresis - Video* &
DNA electrophoresis*
& DNA-electropherogram
2.
DNAg fingerprint*... CSI Miami - how to make one*
a murder case*
a
rape case*
Glowing
gloves & CSI &
DNA
fingerprints in Health &
Society
3. DNA Micro-chip
arrays* -->
chip animation*
monitor gene expression in thousands of genes
& changes DNA microarrays
contain 1,000s of short synthetic ssDNA gene
sequences that are are fabricated onto
a glass slide by high speed robotics akin
to Intel chip making.
By passing
cDNA probes
of a cell's mRNA over slide with ssDNA of all a cell's genes,
the
fluorescently tagged cDNA probes are so easy to
see in slide's wells.
normal vs. cancer
cells* & making DNA microchips

Some Other Practical Applications of DNA
Biotechnology - What's been Done in Medicine...
diseases
often involve changes in gene expression and can be
visulaized in lab.
a. RFLP - Restriction
Fragment Length Polymorphism -
markers may be inherited with disease
what is RFLP*
this
polymorphism is the basis of a genetic test for
the Sickle Cell trait --> DdeI
cuts* +
Genetic Info
Nondiscrimination Act
b. Gene Therapy... idea
is to replace defective genes via microinjection*
allowing recombination to
replace defective gene with newly injected gene.
Vectors*
...using
viral vectors to fix a defective gene
Trials: 1. 1990 clinical
trial a patient with ADA Deficiency -Ashanti DeSilva
video then -
now
2. in 2000 - SCIDs
[severe combined
immunodeficiency - a single gene enzyme
defect].
Clinical
trials in 20 patients triggered 5 cases of
leukemia & 1 death: trials
stopped.
retroviral vector
inserted the repair gene into bone marrow cells genes
incorrectly.
c. CRISPR-Cas9 short animation*
& CRISPR
structure*
& CRISPR-Cuts* & CRISPR Potential*
2020
Nobel & Doudna explains CRISPR
& Treatment of a Patient
(SSA)
anecdote: Story of
Discovery of CRISPR system.
d. Pictorial
Chronology of the Gene... &
Genetic Milestones from
Peas to now...
e.
What is involved in Genetic
Engineering today*:
back
to Bb
next lecture 'Genomics'
.
.
SKIP EVERYTHING BELOW THIS
POINTS....
https://www.youtube.com/watch?v=MNFUf8dqk68
How do cells Control Gene
Expression?
How do we know a gene has been active (turned on) within
cells????
one looks for a gene's product, i.e., an RNA
or a protein
an increase in enzyme activity implies gene
action?
no enzyme activity suggests no gene
action.
but, what about pre-existing inactive enzymes
converting into --> active
forms
ZYMOGENS
- proelastase
-----> elastase
- trypsinogen
-----> trypsin
thus, we have 2 possibilities:
1) pre-existing
inactive enzyme --> active
2) de novo (new) enzyme synthesis by
gene action.

How
do cell control gene expression...
by Turning On/Off Genes*
...
a model: in
Prokaryotes: LACTOSE OPERON - Jacob & Monod (1965 Nobel Prize)
in bacterial DNA, a
cluster of contiguous genes, called an operon,
are transcribed from one promoter making a
single long mRNA
glucose E.
coli (grown on) lactose
NO beta-galactosidase
beta-galacotsidase (gene
expression)
lac ---> glu + gal
OPERON = series of mapable-linked
genes that
can be induced (on) to make several enzymes.
p Rg
crp
p
O Sg1
Sg2
Sg3
p
promoter
region - binds RNA
polymerase [TATAAT box for
RNA polymerase] figure
Rg (I gene) regulator
gene
- LacI gene
makes repressor protein - no lactose is present*
O
operator
site
- DNA
regions that binds repressor protein figure
S
structural
genes
- make enzyme
proteins
but
what if regulator binds lactose*
animation
- Lac Operon* the Lac Operon is
Inducible
 Tryptophan Operon is
off when TRP is present & is this a
Repressible Operon*
Tryptophan Operon is
off when TRP is present & is this a
Repressible Operon*
Control
of Gene Expression... in
EUKARYOTES
Gene Action of the Exome (expressed
genes) has
significantly greater complexity
there are no operons present that map as
genes linked together ,
i.e., no
polycistronic mRNAs.
but regulator proteins, inducers
& repressors are present;
and
there's much more
DNA, - but it's
inside a compartment (nucleus)
have
many more promoters - sites where RNA
polymerase binds
enhancer sequence
- sites where enhancers/transcription factors bind
transcription factors
- proteins that help
transcription
and the individual
genes are not contiguous
with each other, thus no operons
Levels
for eukaryotic controls* - transcriptional,
translational,
post-translational
there are multiple
places for control*
- of whether a gene
makes a protein or not..
Summation - Control
Gene Expression*
&
Differential
Gene Activity animation*
 back
back
Some examples of how Eukaryotic gene expression
controls work:
1. Differential Gene Activity...
is the selective expression of genes as crystallin
gene*
i.e., different cell
types express different genes [liver
vs. lens cell]
role of activators in selective
gene expression
(Differential
Gene Activity*)
ex: Growth
factors & Steroid
Hormones (figure*)
2.
Epigenetic
activation/repression of genome... heritable
changes in phenotype
without alteration of DNA sequence, rather by chemical modification to
DNA or
nucleosome histones (methylation,
acetylation, phosphorylations).
examples: Barr bodies (inactivation of one X
chromosome & imprinting).
Epigenetics
modifications* can carry over to all a cell's
progeny in an organism,
a cell's memory of gene action can be inherited by
progeny cells.
3. Molecular turnover - animation*
--> ½
life mRNA's* & longevity
of some proteins*
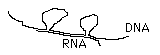
4. Processing of
RNA transcript
(figure*)
cut/spliced in nucleus and capped for transport
intron - pieces
cut out (non gene-proteins)
exons - pieces
transported to cytoplasm
alternative
splicing
= figure 18.11*
5.
Summation -
McGraw-Hill animation on
control of gene expression*AdobeView@Home

Eukaryotic gene expression
controls... examples from cancer:
Cancer often results from gene changes
affecting cell cycle control
proteins:
cancer genes, such as
adenomatous polyposis coli, which cause 15% of
colorectal cancers,
are tumor suppressor genes, a type of Oncogenesg... (an
oncogene is a cell cycle gene
that
has mutated & is now expressed at high levels
turning a normal cell into a tumor cell).
Examples of human cancer Proto-oncogens & Oncogenes:
a presumptive*
& a cancer gene...
Ras a proto-oncogene (a cell division activator)
involved in some 30% of human cancers:
is a G-protein
that promotes other cell division proteins... =
over-expression =
tumors.
a Ras gene mutation -->
hyperactive Ras
protein --> cell division fig
18.24*
p53 a tumor
suppressor geneg is involved in 50% human
cancers:
fig
18.25*
p53 is a transcription
factor that promotes the synthesis
of cell cycle inhibiting
proteins...
thus a p53 mutation
--> leads to excess cell division (cancer tumor).
BRAC1 & BRAC2 are tumor
suppressor genes involved in 50% of
human breast cancers
that when mutated prevents
repair of DNA damage & unregulated cell division.



Genes
and Evolution:
Modern synthetic
Darwinian evolution
predicts that Natural Selection
over time can lead to permanent changes in the the DNA that is inheritable.
but recent studies
have evoked a new science called EPIGENETICS...
study of changes in
gene activity that do not involve alteration to
the genetic code,
but changes that are still are passed to at least one successive
generation.
epigenetics changes the
DNA as a biological response to an environmental
stressor,
changes that can be
inherited through many
generations via epigenetic marks,
but if you remove the stressor epigenetic marks fade & the
DNA code will revert to normal.
somewhat Lamarckian (1744-1829) animals acquire traits
with their life span (giraffe).
ex: 1. drug
geldanamycin produces outgrowth of
Drosophila eyes that can last for 13
generations
no change is DNA sequences and generations 2-13 were
not exposed to drug.
2.
166 fathers who smoked before
age 11 had sons
who had a significantly higher BMI
than control kids of 14,024 fathers.
3. a diet rich in B-vitamins (folic acid & B12 - CH3 donors) fed to pregnant agouti mice
normal pups, but those w/o B-vitamins produces pus
with yellow coats & diabetes.
analogy: the genome is the hardware
and epigenetics is the software:
one can load windows on a Mac; you'll have the same
chip in the Mac (same genome)
but the software will produce a
different outcome - a different cell
type.
end.
 Home |About |News |Syllabus |Lecture-outlines |Links |FAQ |Sitemap |Contact
Home |About |News |Syllabus |Lecture-outlines |Links |FAQ |Sitemap |Contact
MST
II
restriction cuts of normal sickle
beta-gene
( pink is DNA sequence & blue = 4
gel fragments)
_________|__________________|CCTNAGG GAA
_____________|____________
|
|
a
b c
d |
| In
1978, Yuet Wai Kan and Andrees Dozy
of the University of California-San Francisco
showed that the restriction enzyme Mst
II, which cuts normal b globin DNA
at a particular site, but will not recognize and
therefore will not cut DNA that contains the
sickle cell mutation. Mst II recognized
the sequence CCTNAGG (where N = any
nucleotide). Sickle cell disease is due to a
single point mutation in the beta globin
gene on chromosme 11 that changes
CCTGAGG to CCTGTGG. |
MST II restriction cuts of recessive sickle
beta-gene (blue = 3 gel fragments)
_________|___________________CCTGTGG
________________|____________
|
|
a
x
d |
¥
Sickle Cell disease occurs
when the DNA sequence for
glutamic acid is converted to valine. This results from a
change in the nucleotide T
to A. This change
eliminates a site recognized by the restriction enzyme
DdeI.
Restriction enzyme:
DdeI*
(recognition sequence: 5'-C^TNAG-3')
Southern blotting probe:
fragment of ß-globin coding sequence
Pattern result: normal cell = 3
fragments (1 large, a 201bp piece, and a 175bp piece
sickle cell
= 2 fragments (1 large, and a 376bp piece)
fig 20.9*
Thus the number of RFLP piece can indicate presence of
defective alleles.
¥
reading frame
is 1 codon = CAT point mutations at hot
spots - [fig]
mutational
hotspots in bacteria - sequences more susceptible to
change than others
C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T
(MET)
↑ ↑
1st point insertion
or deletion (where
chemical mutagen analogs act during replication)
C-A-T-X-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A =
mutant bacteria
2nd insertion or
deletion
C-A-T-X-Y-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C
= mutant bacteria
3rd insertion or
deletion
C-A-T-X-Y-Z-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T
= normal cells
¥ back
hot spots where mutations occur
with greater frequency.
provides evidence in support of a triplet codon.
|
The index case for AMPD
deficiency was a 18 year old female
with calf pain that revealed a mis-sense mutation at nucleotide 143 in codon 48 of
exon 3 where C
changed to T
resulting in a change of proline
to leucine. This
mutant allele is found in 12% of Caucasians and
19% of African Americans and results in the AMPD deficiency in
muscle biopsies and results
in exercise-induced metabolic myopathy in
humans.
ref: A life Decoded by J. Craig Venter, Viking
Press, 2007, C2, pg 28
back
|
DO
NOT study ANY OF THE material below
Z-DNA
Gene
expressions in
pharmacogenomics
&
toxicogenomics via microarrays
1 cM = about 1 Mb
TRANSPOSONS -
pieces of DNA prone to
moving & creating
repeat sequences
LINE
- long interspersed nuclear
element holds promoter & 2 genes: RT & integrase
an
anomaly - RNA Recoding*
|
|
Simple Tandem Repeats (short- 5n to
6n) or trinucleotide
(3n) repeats can undergo an increase in copy
number by a process of dynamic mutation; # of tandem repeats is unique
to a genetic indiv.
Variation in the length of these repeats is
polymorphic. figure*
individual A has ACA
repeated 65 times @ loci 121, 118, and 129
individual B has a
different repeat pattern at these loci
STR'sa can cause genetic
diseases as well:
CCG trinucleotide occur in fragile sites on human
chromosomes (folate-sensitive group).
fragile X (FRAXA) is responsible for familial mental
retardation.
another FRAXE is responsible for a rarer mild form
of mental retardation.
mutations of AGC repeats give rise to a number of
neurological disorders.
3. Forensics - DNA fingerprinting is the
vogue judicial modus operandi
a
murder case* &
a rape case* + DNA
prints in Health & Society &
DNA Forensic Science
DNA fingerprinting usually looks a 5 RFLP markers
and blood is tested via
Southern Blotting (20.10)
using probes for these alleles
4. Environmental Clean-up...
bacteria can extract heavy metals (Cu, Pb, Ni) from
the environment
& convert them into non-toxic compounds
genetically modified bacteria may be the "miner's" of the future

5. Franken Food... genetically
modified (GM) animals & agricultural crops
Transgenics - organisms
with inserted foreign DNA in their genomes
Animals*
- GFP
novelties* +
Dolly
- animal cloning
companies
---> mammalian cloning
success?
- "pharm" animals (20.18*) --->
transgenic animal movie
sheep carry human blood protein gene
that inhibits enzymes in cystic fibrosis;
artificially insemination, microinjection of
human gene, fertilized ova are put
into a surrogate sheep:
chimeras
mated to produce homozygote- Milk tested for
active protein.
Plants
- genetically modified crop plants
- fig 20.19*
-
to get Ti plasmids in = a DNA gun*
Purdue University Gene Gun movie
- Frankenfood
& Edible Vaccines
- National
Plant Genome Initiative Plan update
future
6.
Synthetic Biology... artificially
manufactured biological systems
- virus models*
(synthetic Biology)
ð
An overview of biotechnology
History
of Biotechnology
Human
Genome Project & Biotech Companies
 HHMI
funded DNA Interactive tutorial
HHMI
funded DNA Interactive tutorial
What
are Introns? and What is the Role of Intron
DNA?
don't
really know, but
Percentage of non-coding DNA during evolution*
goes up.
|
summer & fall
2006: skip this material |
| INTRONS - DNA Junk or sophisticated Genetic Control
Elements? |
 |
 |
 |
Current dogma of Molecular
Biology
DNA --> RNA --> Proteins,
(proteins supposedly
regulate gene expression) figure*
|
in
1977 Phillip
Sharp & Richard Roberts discovered DNA
contains introns
intervening DNA segments that do NOT code for
proteins
a primary RNA transcript is processed by
splicing to assemble protein coding exons
|
Presence
of Introns: Absent in
prokaryotes: they have few non-coding DNA
sequences
as
eukaryotic complexity grows so does non-coding
DNA [figure]
makes
up greater than 95%
of the DNA
less
than 1.5% of
human genome encodes proteins, but all of DNA
is transcribed
40% of human genome
is Transposons
& repeat
genetic elements.
|
Evolutionary
Origins? may have been self-splicing mobile genetic
elements
that inserted themselves into host
genomes
Advent of Spliceosomes:
catalytic RNA/protein complexes
that snip RNAs out of mRNAs,
 would encourage introns to proliferate,
mutate, evolve
would encourage introns to proliferate,
mutate, evolve |
fall 2005 skip ALL OF THIS
MATERIAL
Role of Introns? Not Junk, but
rather Genetic Control Elements
[figure*]
Micro
RNAs - derived from introns? -
occur in plants, animals, & fungi
a) help control timing of developmental processes as
cell proliferation,
apoptosis, and stem cell maintenance
b) help tag chromatin with methyl and acetyl groups
c) may help in alternative splicing
mechanisms
COMPLEXITY:
to build a complex structure one must have bricks & mortar,
as well as an architectural
plan.
DNA, therefore should contain
both - the materials and
the plan:
a) component molecules - proteins, carbs, lipids,
and nucleic acids:
all known living organism use the same bricks and
mortar
b) the difference between Man & Monkey is the architectural plan.
Where is the
Architectural Information? we've
always assumed in the regulatory proteins
Maybe it's in the non-coding mirco-RNAs (intronic
elements)
Thus the greater proportion of the genome of complex
organisms, the introns, isn't junk,
but rather, it is functional
RNA that regulates time dependent complexity?
A Gene Sweep betting pool winner with
closest bet (25,947) was Lee Rowen.

Visualizing Restriction Fragments... to a probe
of a gene (cDNA)
Southern Blottingg
fig*
Sumanas
animation - DNA electrophoresis & blotting*view@home
one can detect specific gene sequence by binding to
labeled probes to DNA fragments
d. 'Human
Microbiome Project'... sequencing human
microbes to correlate with human health
--->
RFLP markers to disease*
& Newer VECTORS - [
animation of translation*view@home]
[
Virus infection
- phage pics - Replication
& role
of tubulins ]
DNA
replication animation quiz*view@home
Review of the Events:
1. DNA pol III
binds at the origin of
replication site in the template strand
2. DNA is unwound by replisome
complex using helicase
& topoisomerase
3. all polymerases require a preexisting DNA
strand (PRIMER) to
start replication,
thus Primase adds a
single short primer to the LEADING
strand
and also adds primers
to the LAGGING strand
4. DNA pol III is a dimer
adding new nucleotides to both strands primers
direction of reading is 3' ---> 5' on template
direction of synthesis of new strand is 5" --->
3'
rate of synthesis is substantial 400 nucleotide/sec
5. DNA pol I removes
primer at 5' end replacing with DNA bases, leaves 3'
hole
6. DNA ligase seals 3'
holes of Okazaki fragments on lagging strand -
cartoon
Review
of the sequence of events
in detail* -
lagging strand and
and
BioFlix anim. of Replication*view@home
DNA Repair mechanisms - 2015 Nobel
Prize in Chemistry*
Linear
DNA replication, Telomeres, and Aging
Sumanas advanced animation
-
de
novo gene origins
current events [Ebola
poster]
DNA
@ 60 Poster & sequencing costs
rmaceuticals... Recombinant
bacteria* = Humulin pen & protropin (an ethical dilemma)
How
the Maize
genome was sequenced by NSF's Plant Genome Inititive
& Forensic
Analyses
e. Genetically
Modified Organisms [1st by FDA
= animation
- Atlantic
Salmon*picture)
2. control
of the Human genome...
How is gene expression
regulated???
SKIP this one page
Micro
RNAs (& siRNA):
function in gene silencing & gene regulation
are single-stranded
RNA molecules of 20-24 nucleotides in
length, that are not translated
into protein
(non-coding RNA) and form short stem-loop
structures that are
partially
complimentary
to mRNAs and that can
down-regulate or possibly activate gene expression.
some 400 miRNAs are
known in the human genome & function as
small interfering RNAs
(siRNA) or miRISC's by hybridizing to
mRNA, forming a dsRNA duplex
& block translation*
found in MODEL
eukaryotic organisms as: roundworms, fruit
flies, mice, humans, & plants (arabidopsis);
ex:
miRNA -->
1) BARR
Body*
& 2) CCD &
3) heart disease
2) TMK-ebola siRNA
Drug
FDA approved Sep 22, 2014
The ability of transfected
synthetic small interfering miRNAs
to suppress the expression of
specific transcripts
is a useful technique to
probe gene function in mammalian cells.
Such non-protein coding RNA
transcripts likely regulate much of the genome via
- miRNA's.
 new roles* for siRNA,
miRNA, and Crispr-gene editing... new roles* for siRNA,
miRNA, and Crispr-gene editing...
C.
Sequencing
Genes
& the
Human Genome Project moon-like
project est: $3 bil & 15 yr
Frederick
Sanger (Cambridge
U.) in May 1975 produced 1st complete sequence of
viral genome
of phage ΦΧ-174
(5,375 np's)
- followed by human mitochondrion with 17K np's.
1st:
methodology - dideoxy*method* & reading* & procedures = sequencing
labs*
used terminator nucleotides for they randomly stop
action of polymerase
when they are incorporated into growing chain marking
its end. dideoxy
video
next:
strategy - shotgun
approach* -
shotgun
video
random DNA
fragments (from a library 500-800n long) are
sequenced using automated
sequencing machines & then ordered relative to
each other via overlap & supercomputing.
next
generation Sequencing
&
Genome Sequencing Standards
timeline:
in 2003 (13thyr) 1st "complete" human genome
sequence was published ($3 billion).
costs: in 2007 Craig Venter's
Genome (dideoxy method) = $7mil ($10
per 1,000 bases).
James Watson's genome (April 2008) =
$1mil (20¢ per 1,000 bases)
CoFactor
Genomics was able to do it next for about
$22,900 (4¢ per 1,000 bases)
EBay
bidder (in 2009) paid $68,000 for sequencing his
genome.
NHGRI
cost estimates - per million bases & per genome bases*
& genome* &
fetal
genome
$1,000 genomes? Novogene's
$850 Sequencing + Your Ancestry DNA
+ 23and me
Sequence
Your Genome
& medical whole genome
analysis &
future
of medicine
Arthur
Kornberg (1918-2007) - 1st to synthesize DNA in test tube
won;
(polynucleotide
phosphorylase)
additional
animations: BioFlix anim. of
Transcription, Translation & Protein Traffic*
Summary
of the Molecular Biology of the Gene*view@home-7
min
Virtual Cell
animation of translation*view@home
and: Life
Cycle of a mRNA
& Nobel Committee on the Central
Dogma
& Virtual
Cell animation of Translation
additional
animations: BioFlix anim. of
Transcription, Translation & Protein Traffic*
Summary
of the Molecular Biology of the Gene*view@home-7
min
Virtual Cell
animation of translation*view@home
and: Life
Cycle of a mRNA
& Nobel Committee on the Central
Dogma
& Virtual
Cell animation of Translation
CRISPR
a gene editing technique* -
What's been done
& medical roles for RNAs...
Luekemia
immunotherapy... inserts genes into
T-cells for cancer receptors (fig*)
3. Single Gene
Expression during development via
RT-PCR-cDNA methodologies
and Differential Gene
Activity ---> fig 20.11*
& DNA chips - normal
vs. cancer cells*view
MST II also
cuts Sickle Cell)
copying
of DNA into DNA seems structurally obvious*???
How is
the transcription of genes
regulatedlater?
DNA
Learning Center video & Eukaryotic
Initiation Complex formation is complex
Roger Kornberg's real-time
animation of transcription (2006 Nobel)*view@home x
& E.M. picture*
&
fig 20.13
How is
the transcription of genes
regulated
.
Long
CRISPR-description + HTR
animation
of their experimental design*
|