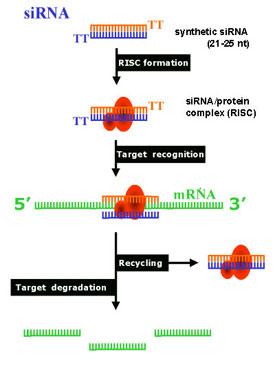
Antisense RNA experiments lead to the discovery of Interference RNA [RNAi]. In 1998, the American scientists Andrew Fire (MIT) and Craig Mello (Harvard) published their discovery of a mechanism that can degrade mRNA made by a specific gene. Subsequently they were awarded the 2006 Nobel Prize in Physiology or Medicine. Small interfering RNAs (siRNAs) are small double stranded RNA molecules containing a "sense" and an "Antisense" strand that by a regulatory mechanism occurring in eukaryotic cells - RNA interference (RNAi) - result in the degradation of a target mRNA in C. elegans. The antisense approach to gene silencing involves injecting an organism with RNA sequence complementary to mRNA transcribed from a target gene. The antisense RNA and sense mRNA hybridize and block translation and the production of an encoded protein. The presence of dsRNA duplex led to what we now recognize as an RNA interference effect.Using genetically altered strains of the roundworm C. elegans (round worms) scientists in fall of 2000, discovered genes responsible for a process called RNA interference (RNAi)—in which double-stranded RNA triggers the natural degradation of a homologous mRNA. RNA interference (RNAi) is a process in which double-stranded RNA triggers the degradation of a homologous messenger RNA (sharing sequence-specific homology to particular "target" mRNAs).Some of the same genes that are responsible for RNA interference appear to also be involved in nonsense-mediated decay, a protective mechanism that may be used by cells to proofread newly created messenger RNA (mRNA) and to prevent the production of defective protein molecules.In an article published in the September issue of the journal Science, Howard Hughes Medical Institute investigator Brenda L. Bass and geneticist Susan Mango and their colleagues at the University of Utah reported that three of seven genes of a gene family called smg were involved in RNAi. Since Bass's discovery molecular biologists have used double-stranded RNAi as a tool to degrade mRNA in cells to shut down the effects of specific genes in C. elegans, Drosophila and many other cell-types. In an experimental test of the role of smg genes they obtained C. elegans strains containing mutations in the various smg genes. They next injected these strains and a wild-type strain with double-stranded RNA designed to interfere with production of myosin, a protein that is critical to muscle development. Thus, they could quantify the effectiveness of RNAi by measuring paralysis in the worms using a crawl assay—a test of the worms' ability to move across a laboratory dish. In the wild-type worms, they found that the progeny treated with RNAi were paralyzed from day one and remained paralyzed.. Therefore the RNAi technique completely interfered with normal muscle development by stopping the action of myosin mRNA. Today RNA interference (RNAi), is a technique in which exogenous, double-stranded RNAs (dsRNAs) that are complimentary to known mRNA's, are introduced into a cell to specifically destroy that particular mRNA, thereby diminishing or abolishing gene expression. The technique has proven effective in Drosophila, Caenorhabditis elegans, plants, and recently, in mammalian cell cultures. To make the technique work in cultured mammalian cells for research purposeds, scientists must deliver small interfering RNAs (siRNAs), which are dsRNAs and of some 21-25 nucleotides (called siRNAs), into the cell. This is done with transfection reagents, which are solutions optimized for allowing DNA and RNA to be absorbed by cultured cells. Thus, RNA interference (RNAi), is a gene-silencing technique used in studying the absence of normal gene action (by disrupting its activity in vivo, i.e., turning it off) in fruit flies, nematodes, and mammalian cells. During the 1990's, scientists realized the power of the RNAi technique to determine gene function by blocking the expression of a specific mRNAs. Sort of a "REVERSE of GENETICS". The introduction of dsRNA mimics a gene knockout of almost any known gene by in effect deletion of the mRNA of the gene. The application of RNAi to mammalian cells has the potential to revolutionize the field of functional genomics. The ability to simply, effectively, and specifically down-regulate the expression of genes in mammalian cells holds enormous scientific, commercial, and therapeutic potential. Artificial siRNAs can be made in the lab by a phage enzyme referred to as DICER. The mechanism involves complexing siRNA into a multi-protein siRNA complex termed RISC for RNA Induced Silencing Complex. Typically, 3-5 double-stranded siRNA molecules are designed per gene in order to find an siRNA that has a strong effect. This means synthesizing and purifying 6-10 RNA oligonucleotides of 20+ nucleotide pairs, at a minimum cost of $1500 per gene. How Does RNAi Lead to Gene Silencing?The basic mechanism of RNAi (siRNA) is thought to be a multi-step process:
The ability of transfected synthetic small interfering siRNAs to suppress the expression of specific transcripts has proved a useful technique to probe gene function in mammalian cells. However, high production costs limit this technology's utility for many laboratories and experimental situations. Recently, several DNA-based plasmid vectors have been developed that direct transcription of small hairpin RNAs, which are processed into functional siRNAs by cellular enzymes. Although these vectors provide certain advantages over chemically synthesized siRNAs, numerous disadvantages remain including merely transient siRNA expression and low and variable transfection efficiency. It is also incredibly difficulty getting cells to take up naked siRNAs. For additional information, please visit "The siRNA User Guide" at Rockerfeller U. Tuschl Lab: siRNA. This site is updated regularly by leaders in the siRNA field as a resource for the research community. Commercial product resource information for siRNA research has been compiled by the Ion Channel Media Group, a targeted biotechnology advertising company at http://www.si-rna.com. Read more: Master of the Cell - The Scientist - Magazine of the Life Sciences http://www.the-scientist.com/article/display/57249/#ixzz0jap9pQ5J http://henge.bio.miami.edu/mallery/150/Charles Mallery, Department of Biology, University of Miami, Coral Gables, FL 33124 Last Update - August 05, 2013
|