| SYNTHETIC BIOLOGY... |
construction
and design of biological system parts or whole organisms
artificially...
Synthetic Biology currently encompasses a number of engineering
strategies including applied
protein design, genome design and
construction, natural product or synthetics drug synthesis,
vaccines, pollution censors, and even the creation of
standardized parts to build circuits into cells.
i.e., the artificial creation of DNA, genes, viri, and cells that mimic,
or surpass, natural systems.
|
beneficial examples: a. synthesize
any gene with 100% accuracy for $1.60 per base pair
b.
design a low cost synthetics drug to treat malaria
c.
build DNA transistors
d.
make a molecular "water wheel" for use in miniature medical pumps
e.
engineer bacteria to do "work"
|
harmful examples: a.
environmental release of an artificial system with harmful
consequences
b.
re-engineering of deadly viri, as smallpox, to cause epidemic

|
|
Specific Virus Examples:
Synthetic Polio Virus
- July 12
,2002
: Molecular Origin of Life
Research or Bioterrorism?
|
|
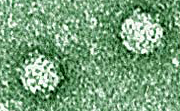
To construct the synthetic polio virus virus, lead scientist
Eckard Wimmer of the State
University of New York at Stony Brook used the poliovirus' widely
known genetic sequence to synthesize that virus
from the building blocks of DNA and a broth of other chemicals.
The followed a recipe they
downloaded from the internet and used
gene sequences from a mail-order supplier. Having constructed the
virus, which appears to be identical to its natural counterpart, the
researchers, from the University of New York at Stony Brook,
injected it into mice to demonstrate that it was active. The animals
were paralyzed and then died.
Wimmer has worked on the genetics and
replication of poliovirus for more than 3 decades. The virus stores
genetic information in a long strand of ribonucleic acid (RNA)
rather than DNA. In the new work, described in
Science (see below),
Wimmer and his colleagues used common laboratory
machines to synthesize DNA strands harboring the same
protein-encoding instructions that a typical poliovirus carries.
|
|
|
Wimmer and his team then mixed the
lab-made DNA with an enzyme that converts DNA into RNA.
 Next, they
added the resulting strands to a mixture of chemicals similar to
that in the cells that poliovirus typically
invades. This brew
generated whole polioviruses that subsequent tests in cells and
animals confirmed as infectious. Next, they
added the resulting strands to a mixture of chemicals similar to
that in the cells that poliovirus typically
invades. This brew
generated whole polioviruses that subsequent tests in cells and
animals confirmed as infectious.
To distinguish any synthesized
viruses from lab contaminants, the investigators introduced subtle
changes into the virus' genetic code that didn't alter the proteins
it encodes. Unexpectedly, however, the newly created viruses turned
out to be much less potent than the typical lab strain. Higher doses
of the synthetic poliovirus were needed to kill mice, for example.
|
|
 |
Cello, J., A.V. Paul, and E. Wimmer.
Chemical synthesis of poliovirus cDNA: Generation of infections
virus
in the absence of natural template. Science. Abstract
available at .
Science OnLine - July 11, 2002
|
|
|
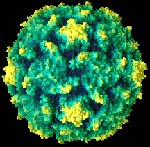
November 2003
Phi X-174 virus synthesized
In
1978 Phi X, a virus that infects
bacteria but is harmless to humans, was the first virus ever sequenced.
By 2004 Craig Venter's (of Genomic sequencing fame) and his research
group (including longtime collaborators Nobel Laureate Hamilton O.
Smith of the Institute for Biological Energy Alternatives (IBEA),
and Clyde A. Hutchinson of the University of North Carolina, Chapel
Hill) at in Rockville, Maryland , in just 14 days, created an artificial
version of
Phi X-174 by
piecing together synthetic DNA ordered from a biotechnology company.
They used a technique called polymerase cycle assembly (PCA) to link
the strands of DNA together.
The sequence and assembly of the synthetic genome matches up
nearly perfectly with the sequenced genome of the natural virus.
Each strand of DNA neatly overlaps its neighboring strands by 20
letters of genetic code. The new virus also infects bacterial cells
just like the natural virus does.
The goal of the IBEA research is to
create a synthetic genome that would be
100 to 1,000 times larger than the phi X
virus. Someday, researchers hope to engineer fleets of
self-replicating microbes that do everything from scrubbing down
emissions at coal plants to producing enough hydrogen to power cars
and trucks run by hydrogen-fuel cells rather than gasoline.

|
|
October 2005
The 1918 Spanish Flu Virus
is Reconstructed
http://www.cdc.gov/od/oc/media/pressrel/r051005.htm
Jeffery K. Taubenberger,
a molecular pathologist at the Armed Forces Institute of Pathology
in Rockville, MD at a CDC
Bio-Safety-Level 3 CDC lab
reconstructed the
genome of the 1918 lethal virus as reported in the
October 7, 2005
issue of Science. They successfully reconstructed the
influenza A (H1N1) virus responsible for the 1918 “Spanish flu”
pandemic (pic + pic). The influenza pandemic of 1918-19 killed an estimated 20
to 50 million people worldwide, many more than the subsequent
pandemics of the 20th century. The Spanish flu virus of the 1918
pandemic was probably a strain that originated in birds. Scientists have found the 1918 virus shares genetic
mutations with the bird flu virus now circulating in Asia.
Taubenberger and his colleagues were able to
piece together the virus's genes from two
unusual sources. One source was fingernail-size
pieces of lung tissue removed at autopsy from
a 21-year-old soldier
who died in 1918 at Fort Jackson in South
Carolina. He was among the pandemic's
675,000 American victims and he provided
intact pieces of viral RNA
that could be analysed and sequenced..
The other source was the frozen body of an
Inuit
woman who died of influenza in November 1918 and
was buried in the permafrost.
The virus's eight "RNA gene
segments" were in pieces. Using gene sequencing
and the polymerase chain reaction they
reassembled the virus.
Two of the
8 genes
are considered to be of greatest importance for
the virulence of the virus: the genes for
hemagglutinin (HA) and
neuraminidase (NA).
Hemagglutinin type 5 [H5] and Neuraminidase type 1
[N1] are protein surface coatings of
the virus. Hemagglutinin A is a
glycoprotein that
binds the virus to the host cell. There are at
least 16 different HA antigens.
Neuraminidase is
an antigenic glycoprotein enzyme found on the
surface of the flu virus. Nine neuraminidase
subtypes are known, many occurring only in various
species of ducks and chickens. Subtypes N1 and
N2 have been positively linked to epidemics in
man. The neuraminidases aid in the efficiency of
virus release from infected cells.
The reconstruction
of the 1918 virus is expected to provide
insights that are immediately useful to the
virologists and epidemiologists charting the
flow of hundreds of flu strains through dozens
of species. Researchers hope to
identify
the mutations that are necessary for adaptation
to a human host. It will be especially
useful to compare the
similarities
between the Spanish flu virus 1918
virus gene sequence
to the H5N1 of the
current "Avian Flu", as well as
to those of the "Asian flu", which emerged in
1957, and the "Hong Kong flu", which circled the
globe in 1968. Hybrid viruses containing genetic
features of each in different combinations can
then be constructed and studied in the
laboratory.
The biological
properties that confer virulence to pandemic
influenza viruses are poorly understood.
Evaluating the biological properties of the
individual genes that make up this virus (CDC
has examined four of the virus' eight genes so
far). Scientists will continue to study the hemagglutinin (HA) gene to better understand the
role of HA in causing inflammation of the lung,
a factor that may contribute to the overall
increased levels of illness and death associated
with this virus.
The strain of avian influenza that has led to the
deaths of 140 million birds and 60 people in
Asia in the past two years appears to be slowly
acquiring the genetic changes characteristic of
the "Spanish flu" virus that killed 50 million
people nearly a century ago.
How far
"bird flu" has traveled down the evolutionary
path to becoming a pandemic virus is unknown.
Nor is it certain the worrisome strain,
designated influenza A-H5N1, will ever acquire
all the genetic features necessary for rapid,
worldwide pandemic spread.

|
|
|
|
A
bacterial example of Synthetic Biology
J. Craig Venter,
a principle investigator (P.I.) of the
Human Genome Project
is attempting
to make a
synthetic new type of bacterium using DNA manufactured in the lab; using the sequenced the genes
of a bacterium called
Mycoplasma
genitalium, a gram-positive parasitic bacterium, whose primary infection site
may be the human urogenital tract. It probably causes
non-gonococcal urethritis.
The complete nucleotide sequence is
580,073 base pairs, the smallest known genome of any free-living
organism, has been determined. A total of only
517
genes (480
encoding for proteins; the rest for RNAs) were identified
that included genes required for DNA replication, transcription and
translation, DNA repair, cellular transport, and energy metabolism.
> researchers began
systematically removing genes to
determine how many genes
are
essential for life. In 1999, they published the
narrowed the needs of M. genitalium
to
between 265 &
350 genes.
> How many genes
does it take to make an organism? What is the minimum genes a cell needs?
The scientists at The Institute for Genomic Research (TIGR)
who determined the Mycoplasma
genitalium
sequence followed this work by systematically destroying its genes
(by mutating them
with
insertions) to see which ones are essential to life and which
are dispensable. Of the 480
protein-encoding genes, they
conclude that only 265–350 of them are
essential to life.
>
The next step
is to artificially assemble these 300+ genes to create a
synthetic cell.
 |
|
| |
| |
| |
| |
 Next, they
added the resulting strands to a mixture of chemicals similar to
that in the cells that poliovirus typically
invades. This brew
generated whole polioviruses that subsequent tests in cells and
animals confirmed as infectious.
Next, they
added the resulting strands to a mixture of chemicals similar to
that in the cells that poliovirus typically
invades. This brew
generated whole polioviruses that subsequent tests in cells and
animals confirmed as infectious.
