Only 10% of cells in the body are human; the vast majority are bacteria, viruses, and other microbes of many species on your skin and in your gut. Humans live in a symbiotic relationship. At birth , babies emerge from a sterile environment, but an infant's intestines then rapidly become home to one of the densest population of bacteria on Earth. Most are harmless, some very helpful, and a minority are pathogenic and can cause diseases.
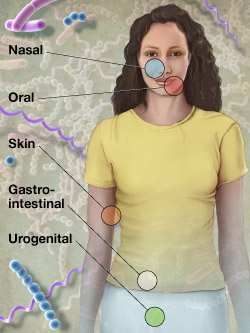
The NIH in December of 2007 launched the HMP with $115 million for awards to researchers over the next five years. It is modeled after the Human Genome Project. The first 35 awards were made in summer of 2009. Initially, researchers will sequence 600 microbial genomes, completing a collection that will total some 1,000 microbial genomes and providing a resource for investigators interested in exploring the human microbiome. Next, they will continue with metagenomic analysis to characterize the complexity of microbial communities at individual body sites, and to determine whether there is a core microbiome at each site using 16S rRNA sequencing and metagenomic sequencing. Several body sites will be studied, including the gastrointestinal and female urogenital tracts, oral cavity, naso-pharyngeal tract, and skin. The most popular site for human skin microbe is the forearm, which is home to 44 different species. The most barren region is behind the ear, with only 15 species typically. Different ecosystems of bacteria are associated with different maladies. Bacteria causing psoriasis favor the outer elbow, while eczema bacteria prefer the inner elbow.
The goal of the project is to identify which microbes are harmful and to figure out ways to prevent or treat disease. For example: Matthias Tschop, of U. Cincinnati College of Medicine, identified 383 microbial genes that differed significantly between pairs of slender and obese twins.
1. characterize microbes of mucosal surfaces
2. correlate microbes & associated diseases