|
Proteins
are more commonly Classified by FUNCTION
'rather than by
structure'
| Enzymes |
catalytic
activity A
------> B |
| Transport
Proteins |
bind &
transport ligand
molecules (hemoglobins) |
| Storage Proteins |
ovalbumin (egg), ferretin
(iron),
casein
(milk) |
| Defensive Proteins* |
provide
protection: antibodies
(IgG),
fibrinogen,
thrombin,
and snake venoms
(digestive enzymes) |
| Contractile
Proteins |
can
contract, change shape (actin
& myosins)
and
make up elements of cytoskeleton &
muscles |
| Structural
Proteins |
provide support... collagen fibers of tendons (wounds),
elastin of ligaments, keratin
of hair & feathers,
fibroin
of silk & spider webs |
| Hormonal-signal
proteins |
coordination of
physiological activities (insulin) |
| Receptor Proteins* |
respond to
chemical stimuli: includes
receptors, transcription factors
& enhancers
|
|
nomenclature |
early
protein naming was based on solubility |
|

|
animation
about protein functions*view@home |

|
Structure and Properties of
all functional Proteins
|
PROTEINS - are POLYMERs of AMINO ACIDS with
biological reactivity...
N-gly-asp-leu-his-met-pro-phe-trp-tyr-lys-ser-val-C
N--G---D----L---H---M---P----F---W--Y--K---U---V--C
made of alpha-L-amino acids (commonly just
20 occur in cellular
proteins)
of the 20 of the common
amino acids, how many amino acids
do you think Humans can synthesize*
??? all, half, none.
|
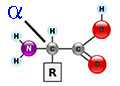
STRUCTURE of Amino
Acids
alpha amino
acid* |
|
|
an amino
acid is a ZWITTERION...
a molecule with 2 functional groups of
opposite charges,
aa's contain 2 functional
groups: a
carboxyl group (-COOH) &
an amino group (-NH2)
i.e., a weak base (NH2 = NH3+)
and a weak acid (COOH = COO-).
& each of these functional groups are
often ionized* at
physiological pH
[weak acid:
a proton (H+) donor
& weak base: a
proton (H+) acceptor].
Isoelectric Point - a pH where there is
no net charge
in a molecule. [NH3+]
& [COO-]
pK - the pH at which all the groups
are 50% ionized &
50% non-ionized |
 What
is charge of glutamic
acid at pH
3.0
?
at pH 9.0 ?
(ans*)
What
is charge of glutamic
acid at pH
3.0
?
at pH 9.0 ?
(ans*) |
| What is an
alpha amino
acid? |
 |
NH2-group is bound to an asymmetric alpha carbon:
--> αlpha = 2HN-C-COOH - βeta = 2HN-C-C-COOH - γ-gamma = 2HN-C-C-C-COOH
β^
γ^
20
different chemical (R) groups
make the common aa's
& |
|
these 20 ubiquitous & universal amino
acids are found in all living systems. |
|
Remember amino acids are
optical isomers: D or L
|
Every amino acid (but one - glycine)
exists in two optical isomeric forms,
each with different arrangements of atoms in
space. |
i.e., 2 optical isomers
are mirror images of each
other
|
► but only the α-L-optical
isomer aa's
occur in biological proteins
... an anomaly of molecular
evolution? |
 |
the properties of the various
proteins depends on the chemical properties of
the amino acids
amino acid's are CLASSIFIED
by the CHEMICAL PROPERTIES of R or Side
groups
attached to α-carbon, which
defines its chemical & physical
properties. |
Polar Charged
ACIDIC
Polar Charged
BASIC |
negatively
charged amino acids - ASP
& GLU
R
group contains a 2nd COOH that
ionizes above pH 7.0
|
positively
charged amino acids - LYS,
ARG, HIS
R
group with
a 2nd NH2
that protonates below pH 7.0
figure* |
| Polar
uncharged |
includes SER, THR, CYS, TYR,
ASN, GLN
are soluble in water, i.e., hydrophilic (attract H-bonds)
figure*
contain hydroxyl or amino
functional groups
|
non-polar
(aliphatic) |
includes GLY, ALA, VAL, LEU, ILE,
MET, PHE, TRP, PRO,
figure*
all contain only hydrocarbons
R groups
= hydrophobic
like lipids |
| AROMATIC |
(hydrophobic
non-polars) PHE & TRP
(TYR)
& MET, CYS
all contain R groups with ring
structures*
or
Sulfur* |
 Table of the Amino Acids*
Table of the Amino Acids* |
| peptide
bond |
COVALENT LINK between carboxyl
end (COOH) of aa-1
& amino end (NH2) of
aa-2 = Peptide Bond |
|
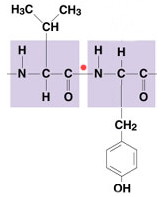
|
forms a dipeptide* or eventually a polypeptide* |
| formed by a condensation reaction
(removes water
= dehydration synthesis) |
|
peptide bond is
intermediate in length
& strength between
C-C & C=C |
|
thus a peptide bond is shorter
& stronger than
C-C and |
|
longer, but weaker, than C=C, however it acts like one,
thus |
|
there is no free rotation
(attached group in same
plane) figure* |
|
and the peptide bond is Flat and Planar* (with
R-groups trans to each other) |
non-protein examples of peptide bonds:
Capsaicin (chili peppers active ingredient)
NutraSweet (dipeptide: L-aspartyl-phenylalanyl methyl
ester) |
|

|
Review animation
of Functional Groups in a tripeptide*view@home
|
PROTEIN STRUCTURE
- Variety of a
linear polymer of amino acid sequences is
infinite...
|
- A protein of 100
amino acids made with the 20 different
known amino acids
can have 20n
(n = protein
length = ~) different linear
sequences for a protein
|
- Average
protein has 300 to 400 aa's
& has a MW
of 30,000 to 45,000d
|
- many
often have a globular
(spherical*)
3-D
shape & are negatively charged
|
|
|
- some ≈ 1,000 proteins
have been chemically sequenced (now
done by gene data bases)
|
- E. coli (a human
intestinal bacteria) makes about 4,250
proteins
|
- Human
Proteome Project has mapped
with an estimated 30,057
human proteins
made from only 17K protein
coding genes (~3% of genome).
|
 |
| 4 levels of protein structure are
recognized |
|
primary
sequence |
linear
sequence of aa's from N-terminal to C-terminal |
| NCC-NCC-NCC-NCC-NCC-NCC-NCC-NCC-NCC
figure* |
|
|
|
secondary
structure |
regular,
recurring orientations of aa's within a
peptide chain
due to H-bonds forming alpha helix
& beta
sheets figure*
|
|
|
|
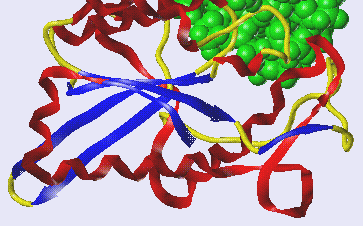
tertiary
structure |
3D -
conformational shape due
to
weak electrostatic
interactions
with other molecules
make the shape of many soluble enzymes
globular figure* |
|
|
 quaternary structure
quaternary structure |
2 or more
polypeptides or protein
subunits interacting to give
to give a unique 3D spatial shape to a
protein figure* |
Animation of the
Structure of Proteins*view@home
PRIMARY SEQUENCE - ex: primary sequence
amination*view@home
& Transthyretin*
|
Some
properties of
proteins due to Primary Sequences... |
| Polymorphism |
some proteins
may vary in their
primary amino acid
sequence,
but can have the same biological
catalytic activity
ex:
catalase/peroxidase enzymes
2 H2O2
--> 2 H2O
+ O2
inter-specific : between
species - different aa sequences (cow vs. human)
intra-specific : within a species
(liver vs. kidney) |
| Invariants |
primary
sequences that do not vary significantly.
ex: ubiquitin* &
histones*
ubiquitin (universal to
eukaryotic cells) signals a proteins
degradation
histones - alkaline
protein that bind with DNA |
Site
Specificity |
some aa sequences
determine intra-cellular location for a
protein & activities:
Signal Sequence*, prosthetic binding sites*,
etc... |
Homologous
Proteins
 |
Proteins related by common
evolutionary history...
i.e., proteins which are derived
from common ancestor,
can occur in different species, or same
species (if 1 gene
duplicated)...
may perform similar cellular
functions in the different species.
ex: Hb* & cytochrome-C*
in ducks &
chickens
= 2 variants
in yeast &
horses
= 48 variants |
Secondary Level - a regular recurring
pattern/shape in proteins due to H-bonds
| alpha
Helix... |
animation* :the peptide backbone is wound
around a long axis core* |
|

|
forms a rigid cylinder*
- a right handed helix
- (counterclockwise) |
|
R-groups of
amino acids radiate outward* |
3.6 aa per 360o turn
- 1.5 Ao/residue
(5.4 Ao
per turn)
1 angstrom = 1 x 10-10
meters (or
0.1 nm) |
diameter of helix is
1.2nm - same
diameter as DNA major groove,
thus a-helices are common in DNA binding proteins. |
alpha helix is formed naturally by H-bonds*
H of
N (of one aa)
& -C=O
(of 4th aa down) |
helix is
typically 10-15
residues long,
⅓
of a typical protein is in alpha helices
and ⅓
is beta sheet |
|
|
Beta sheet...

|
a linear
extended ZIG-ZAG* pleated sheet
conformation |
| formed by
H-bonds... intra- & inter-chain figure* beta sheets* |
| beta sheets are
usually 3 to 10
residues long |

| ENZYMEs... are proteins
responsible for catalysis*
(term is from Gk -
"in leaven") |
1st described in 1880's by Louis Pasteur
and
called "ferments"
(their ability to ferment of sugars)
1st
enzyme crystallized
in 1926
by Sumner - was Urease
breaks down urea* into NH3 & CO2 |
What is it that enzymes actually
do?
enzymes regulate
metabolic reaction
rates in cells,
i.e., they control
metabolism
"are
molecules that
accelerate
catalytic
chemical reactions [ A ---> B
] in cells,
by breaking
covalent bonds and/or forming new
covalent bonds".
(most
cellular catalysis is by proteins -
? but
why not all?) |
they're biological
CATALYSTS*... but, differ
from classical chemical (metal-Fe) catalysts in
that:
1. have complex specific
structures (a unique primary
sequence of aa's = unique 3D shape)
2. act only upon a
specific substance (a
substrate)
3. do not change the
direction (fig 8.14 energetics*) of a chemical reaction
4. enzymes
work by lowering Ea*
bringing reactants
together & thus speeding up* reaction
rates*
|

|
| Some Terminology & Properties of Enzymology: |
besides a protein
part, many enzymes may require a cofactor or coenzyme*...
i.e., protein
holds a coenzyme
in proper orientation to catalyze "bond breaking - forming" |
|
cofactor
|
small inorganic ions that
help catalyze electron [redox] reactions
Cu+2
; Mg+2
; Mn+2 ;
Fe+2
etc... |
|
coenzyme
|
organic,
non-protein molecules, which catalyze
[redox] reactions
many are vitamins
such as NAD+* (or vitamin-B3),
that...
gains/loses e- ;
transfer groups
; break
bonds, etc... |
|
reaction
path |
E + S <---> ES
<---> E +
P
catalytic
cycle of an enzyme
ex: sucrase reaction*(fig 5.16) |
|
active
site |
portion
of enzyme protein that attaches to the
substrate
by means of weak chemical
bonds* to lower
Ea...
(via
H-bonds, ionic bonds, hydrophobic forces,
etc...)
some
theories on the 3D-active site SHAPE*... |
 end 8
end 8
|
Structure of ENZYME-SUBSTRATE
Complex:
√ enzymes are proteins with a specific 3-D shape,
that
binds substrate to its ACTIVE
SITE
lock & key*
vs. induced fit*
√ shape of enzyme is critical to its ability
to convert A
---> B
change an enzyme's 3D shape & it won't
bind substrate
►
What will change shape of a protein [active site] & can thus lead to
denaturation*?
Cells regulate pH and tmperature via homeostasis,
thus changes in...
1. temp -
increases kinetic motion, breaks H-bonds
2. pH -
changes ionic charges, can alter shape
3. inhibitors
- chemicals that bind to enzyme
& change its activity
[competitive/non-competitive inhibition]
4. poisons -
organo-phosphorous compounds (many insecticides)
bind to enzymes of nervous system & thus kill
► Example of a Mechanism of Action
of an enzymr's Active Site - Serine Proteases*

|
Characteristic Graphical Curves of
Enzyme Activity |
or
how to determine if a cellular reaction A —> B is enzymatic
???
by analyzing the
kinetics of the reaction. |
| |
1. Rate
Vs. [
E ]* is
a linear line,
(same as metal catalyst)
(0.8
ml O2/min)
|
| |
|
2. Rate
Vs.
pH
reveals an optimum |
|
figure* |
|
3. Rate
Vs.
Temperature
reveals an optimum |
| |
4. Rate
Vs.
[ S ]*
shows a saturation curve
most definitive curve describing enzyme
activity |
 |
| MICHAELIS-MENTEN Enzyme
Curve
[a Graphical Plot of
enzyme activity] |
a graphical plot
of rate (amount of product per unit
time*) vs. [ S ]
shows the curve saturates*
... reaches a point where [S] equals maximum velocity
of rx = Vmax ◄ |
the Enzyme
Curve defines a new constant relative to
many enzymes:
►
Km
is the
substrate
concentration
at which the rate is
one-half [½] the
maximal velocity (Vmax)
(in the Michaelis Menten
Enzyme curve* --> Km = 0.08
mM)
it's the amount of [S] needed to
reach ½Vmax
it's a "rough" measure of affinity of enzyme
for its substrate |
Compare 2
enzymes*:
G-6-phosphatase [Km = 1 mg] & G-1-phosphatase [Km = 26 mg]
it takes
much less substrate for G-6 to reach same
kinetic point
thus G-6-phosphatase is more
efficient than G-1-phosphatase.
|
Use of M & M
Curves - especially with
environmental inhibitors Enzyme Inhibition*
competitive
inhibition... binds to active site
...reversible 'raises' Km
same Vmax
noncompetitive
inhibition...
binds to an allosteric site
same
Km
lower Vmax
|
 |
| Plot of rate (amount of
product per unit time) vs [S] at constant [enzyme] |
|
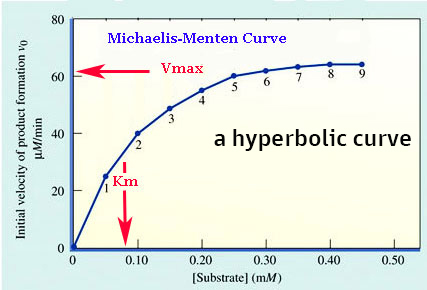
|
BACK
figure of
enzyme saturation*
|
1. SATURATES... at high [S] = Vmax (maximum velocity) in
graph
Vmax = 62
uM per min @
a constant [E]
2. Km = Michaelis
Constant is substrate concentration
when the rate is
one-half maximal velocity (Vmax)
in
graph above
Km = 0.08 mM of substrate
3. Km
can indicate a measure of affinity of an
enzyme for a
substrate;
it is amount of [S] needed to reach
½
Vmax
|
Enzyme Inhibition - is a reduction in
enzyme activity due to action of
a non-substrate molecule
on an enzyme's catalytic activity [shape = binding]..
Irreversible*
inhibitor
covalently binds to enzyme permanently
inactivating it
Competitive*
inhibitor binds to active site, because it looks like
substrate
animation
...is reversible; can be removed by excess
substrate
i.e., substrate can out compete inhibitor for active
site
...it raises the Km value, but has same
Vmax + an ex: Viagra
Noncompetitive* inhibitor binds to an allosteric
site, [not
active site]
animation
...isn't reversible;
...thus inhibitor removes a fixed amount of enzyme -
lowers Vmax
...it has same Km, but
lowers the Vmax
Regulation of Metabolism via Enzyme Cooperativity & Allosteric
Regulation
regulates
enzyme activity by changing
protein shape
conformations
ex: Feedback inhibition* an end product inhibits an
initial pathway enzyme
animation
by altering
efficiency of enzyme action.
many enzymes function by allosteric
sites and
regulation by active
vs. inactive forms*
 summation of
Michaelis-Menten
enzyme Regulations*
summation of
Michaelis-Menten
enzyme Regulations*
MAJOR ENZYME CLASSES...
[ Enzyme Nomenclature Database ]
1. Oxidoreductases
[dehydrogenases] |
catalyze oxidation
reduction reactions, often w coe NAD+/FAD
Alcohol dehydrogenase
[EC 1.1.1.1]
ethanol +
NAD+ -----> acetaldehyde +
NADH |
| 2. Transferases |
catalyze the transfer of
functional groups
Glucokinase (hexokinase)
[EC 2.7.1.2]
glucose +
ATP -----> glucose-6-phosphate + ADP |
3. Hydrolases
|
catalyze
hydrolysis - adds water across bonds
as C-C
Carboxypeptidase
A
[EC 3.4.17.1]
[aa-aa]n + H2O
-----> [aa-aa] n-1 + aa |
| 4. Lyases |
add or remove functional
groups to C=C bonds
Pyruvate decarboxylase
[EC 4.1.1.1]
Pyruvate
-----> acetaldehyde + CO2 |
5. Isomerases
[mutases] |
catalyze isomerizations
- change from one isomer to another
Maleate isomerase
[EC 5.2.1.1]
maleate
-----> fumarate
(cis-trans isomerization) |
| 6. Ligases
|
condensation of 2
substrates (often with splitting
of ATP)
Pyruvate carboxylase
[EC 6.4.1.1]
pyruvate +
CO2 + ATP ----->
Oxaloacetic acid + ADP + P |
7. Translocases
|
catalyze the movement
of molecules across membranes
(ATP)
Phosphate
transporter
[EC 7.3.2.1] |
|
*Common names of
enzymes and their cellular roles* |
 <-- next presentation
<-- next presentation
SKIP ALL THE PRESENTATION MATERIAL BELOW
Electrophoresis - passage of
charged protein mixture through a porous
medium (gel) via its
electrical charge that results in
separation
of proteins by size
& charge* |
|
PAGE |
polyacrylamide gel electrophoresis = PAGE* in gel chambers & gels
protein
fingerprints*
can
indicate evolutionary relations
|
|
SDS-electrophoresis
|
Sodium
dodecyl sulphate [SDS] -
separates proteins via their mass
(MW)
SDS binds to peptide
bonds linear-izing a protein with (-)
charges
Fig1* &
animation of SDSview@home
|
|
isoelectric
focusing
|
proteins migrate thru pH gradient gels to
their isoelectric
point*,
i.e., pH where no
net charge & then stop migrating |
|
2-D electrophoresis
|
combines
isoelectric
focusing & SDS-electrophoresis
procedures*
and
a sample
compares leukemic cells*
|
 |
Protein
Identification &
Quantification |
colorimetric reaction [Biuret
& Bradford]
where amount of
color produced is proportional
to amount protein present...
"3
most cited science papers are on methods
to measure proteins"
[ create a
Standard*Curve* ]
|
back
next lec
copyright
c2022 Last update
- May 2022
Charles
Mallery, Biology
150, Department of Biology, U. of
Miami, Coral Gables, FL 33124
|