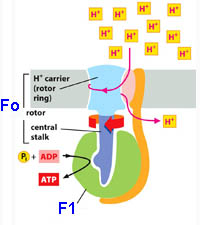
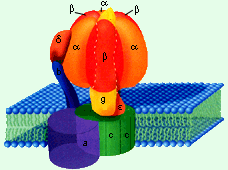
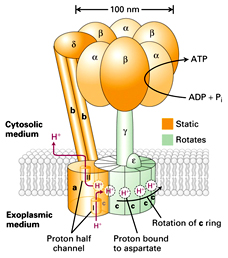
*
*outer membrane contains - porin* - a channel protein
that allows diffusion molecules ≈ < 5,000 daltons
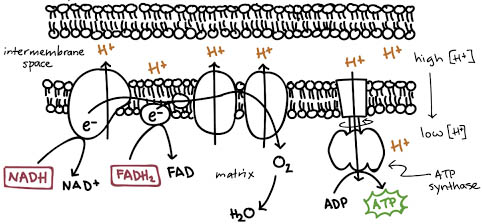
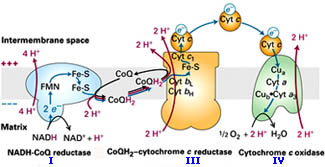
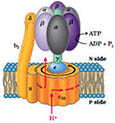
inner membranes
- IMPERMEABLE to most
molecules, esp. to H+
mitochondria
& ecb 14.8
pg461*
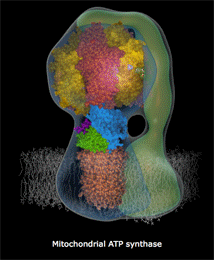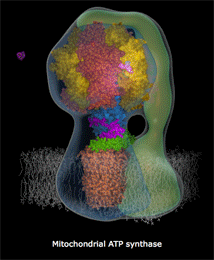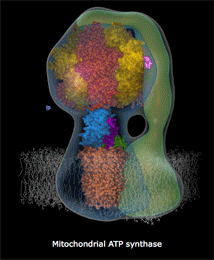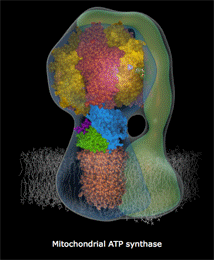
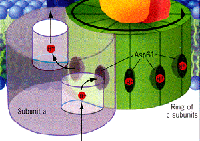
& mitochondrial
tomographyviewed earlier
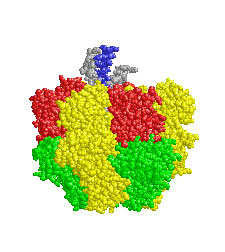
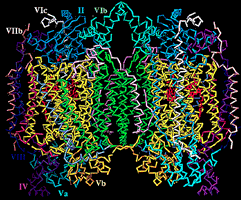
inner membranes -
some 70% protein & 30% lipid... & its
components*
include: