|

|
ENZYMOLOGY - Enzyme… from the
Greek ένζυμο, énsymo,
which means
én
("in") and
simo
("leaven")
-
enzymes are catalytic proteins that ACCELERATE
REACTION RATES
- i.e.,
they control
metabolism
molecules (mostly protein
vs. ribozymes)
that catalyze
chemical reactions (A--->B) in cells by
breaking
old covalent bonds & forming new
covalent bonds
-
are
biological catalysts… but,
different from a chemical catalyst -
have complex structure
(sequence of aa’s) that act only upon
specific substrates
& don't change direction (energetics)
of rx.
catalysis* = acceleration of rate
of a chemical reaction via a catalyst
|
|
|
- enzymes convert
substrates to products w/o themselves changing
|
|
ex: cAMP
protein kinase A* [2.7.11.11]
group
of enzymes that transfer P from ATP to SER on
proteins
|

|
|
Some important early dates in Enzyme History
1833 Payen &
Peroz - alcohol
precipitate of malt barley holds heat
labile
components that
converted starch to sugars (1st enzyme proteins)
1878 Wilhelm
Kuhne - coins term
'enzyme' : Greek "in leaven"
1897
Hans & Eduard
Buchner
- yeast
'zymase' + ferment sugars = CO2
& ETOH
1898 Emile
Ducleaux - customizes
the use of suffix "ASE " for enzyme naming
1900 E.
Fischer - describes
the stereospecificty of
enzymes (lock & key hypothesis).
|
1st enzyme crystallized urease
[EC
3.5.1.5], 1926
James Sumner
Sumner
was first to purify a protein fraction
with catalytic activity...
2 NH2-CO-NH2
+
2 H2O
-----> 4 NH+4
+
2 CO2
the purity
of an isolated enzyme is based upon its
crystallization... |
|
- 1,000s of enzymes
have since been purified &
crystallized
-
except for ribozymes (that also have catalytic
activity) - all are proteins
- proof
that a biological activity is due to an
enzyme has usually been...
noting if the loss of biological
activity
that is result of proteolytic digestion. |
|

|
|
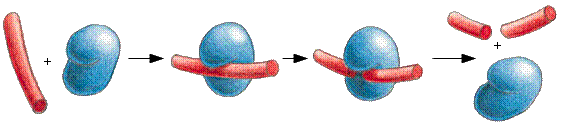
ENZYME
REACTION PATH
E + S
<--> [ES]
<--> E + P
enzymes catalyze
reactions by
lowering the energy
of activation... Ea
ecb3.12* hexokinase*
via a
dissociable complex fig
|

|
There is no difference in
free energy between an
enzyme catalyzed reaction
and an uncatalyzed reaction,
but a non-enzyme catalyzed
reaction requires higher
initial energy input than an
enzyme catalyzed reaction.
|
Some Terminology
substrate, product,
enzyme... self explanatory
|
Rubisco

|
Cofactors* : non-protein
compounds, required for a protein's
biological activity... often as small inorganic
ions...
including many metal ions: Cu, Mg,
Mn, Fe [Fe-S proteins],
which act
as activators
&/or inhibitors of activity...
Table of
vitamin cofactors:
Coenzymes*
: small non-protein ligands
that catalyze
reactions…
+/-
electrons, transfer a
group,
forms or breaks a
covalent bond, etc...
NAD+*
(NADP+) :
redox coenzymes - dehydrogenations*;
H+
carrier and/or
electron transfer:
1st proteinaceious
reactions were
likely e- transfers?
FAD*
:
another redox coenzyme
CoASH*
:
acyl carrier(3HC-C=O) via
sulfhydryl (-SH)
LIPOIC acid : oxidative
de-COOH of alpha-keto acid
prosthetic group:
large complex organic molecule,
which may have catalytic activity
(heme)
a Redox reaction :
Succinate
dehydrogenase example
|
| |
|
Detailed Active Site
Mechanisms of Enzyme Action:
3 examples...
the chemical reaction
scheme by which an enzyme acts
upon its substrate...
1.
Lysozyme example: (2013
Nobel Prize Chemistry for Computer
Modeling Molecular Processes)
- an
enzyme
that
cuts polysaccharide* glycosidic
bonds by hydrolysis (adds H2O)
- active
site is a long groove, holding six
sugar units...
has 2 acidic side side
chains (GLU & ASP) hold
substrate
- breaks
glycosidic bond (…
-C-O-C- …)
via bond
strain & distortion of glu &
asp
- enzyme binding of
substrate, bends bonds from a stable
state, lowering Ea.
acidic
side group of
GLU
provides a
proton to attack glycosidic bond
[pic],
and ASP favors
hydrolysis of glycosidic bond [lysozyme
animation* & ecb 4.35*].
2. Protease*
hydrolysis
of peptide bonds: serine protease*
catalytic
sites hold ser195, asp102,
& his57: -OH of ser195 e's attack C=O of
peptide bond & transition state is
held by H-bonds;
e's break peptide bond & release
part of protein; H-O-H is split &
added to split bond.
3. catalytic
action of cAMP
dependent Protein Kinase A* - e's of ATP are delocalized by
LYS & Mg+2;
new bond forms between SER-OH &
γP; bond between
βP-γP broken =
ADP + P-protein.
"a
proper shape of an enzyme is critical
to its ability to catalyze a
reaction".
|
 How
do we determine the rates of a enzymatic
reaction A
--> B
??? How
do we determine the rates of a enzymatic
reaction A
--> B
??? |
Enzyme
Kinetics…
define the physical & chemical properties of
enzyme by mathematical and/or graphical
expression of the
reaction
rates of enzyme catalyzed
reactions.

|
Catalase
EC
1.11.1.6 |
Dye + H2O2
---> 2 H2O
+ colored dye
its reactivity is made
visible using a marker that, when
oxidized by enzyme using H2O2
as the oxidizing agent, yields a color
change detectable by spectrophotometric
methods. |
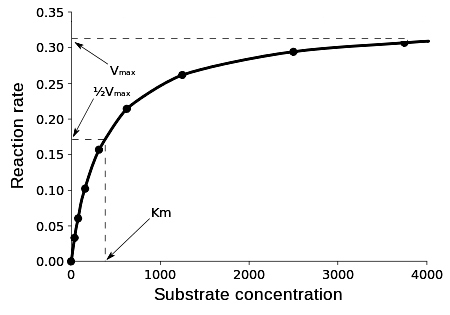
Characteristic
Graphical Enzyme Kinetic Curves:
this is how to determine if the
reaction
A —> B is
enzymatic???

Enzyme rate (v) substrate
concentration curves:
|
A
Michaelis-Menten Curve
|
v vs. [S] curve
defines a rectangular
hyperbola
|
 |
at low [S], rate is
directly proportional to [S]
|
| at higher
[S],
rate declines giving a hyperbolic
curve |
|
one
gathers data points for an enzyme
curve -->
-
ecb
3.27*
|
1st & 2nd order reaction kinetics
alone are NOT sufficient
to describe the shape of a
plot of enzyme reactions (above)
|
A
<--k1-->
B for
1st order
Rx
dP/dt
= k1 [A]
= linear
response thus no |
| A
+ B <--k2-->
C for
2nd order
Rx
dP/dt
= k2 [A]
[B] = also linear
thus no |
| |
 |
in
1913 Leonor
MICHAELIS
&
Maud MENTEN
proposed a mathematical modeling of
enzyme reactions using algebraic
expressions and rate
constants to define
a rectangular hyperbola response.
k = rate / [A] *
[B]
|
|
k1 k3
E + S <---------> ES <--------> E + P
k2
k4
|
|
|
Some assumptions
in the Michaelis & Meneten
enzyme equation derivation...
1)
rate formation ES
complex from E
+ P is
negligible, i.e., can ignore the
rate constant
k4
2)
rate LIMITING step is disassociation
of ES to
E + P
= k3
(speed of
dissociation)
k3 (rate constant) is #
of molecules converted by this
reaction per unit time
v =
(dP/dt) = k3 [ES]
3) an important
state of the ENZYME is termed
FREE ENZYME which
is able to react
bound
enzyme
=
[ES]
free enzyme
=
[Et
- ES]
total enzyme = [Et]
= [Et
- ES] +
[ES]
|
|

|
"Die
Kinetik der Invertinwirkung" Michaelis,
L.; and Menten, M.L. (1913) Biochem. Z.
49, 333-369. |
Derivation of
Michaelis-Menten Enzyme Kinetics...
|
their
derivation of equation occurs at a time
when...
the rate
of formation of ES complex is
equal to rate of destruction
(break down),
i.e., at a time when [S] >>>>
[E],
so that total E is
bound in ES
complex and
thus reaction
works like a 1st order reaction enzyme
catalyzed reaction
|
the rate limiting equation thus becomes
destruction of ES... v = (dP/dt) = k3 [ES]
|
it would be easy if we could measure the
concentration
of [ES]...
say in a spectrophotometer
but, its
presence is fleeting....
so then the real function
of of M&M kinetics is to be able
to express
[ES]
in terms of E
& S
alone, which are
measurable quantities…
measuring
[ES] quantitatively
is very difficult [ a stopped
flow apparatus ecb
3.28* ] |
the derived
M & M equation
is then :
v =
Vmax [S] --> gives graph*
Km +
[S]
mathematical
derivationtake-a-look
y =
a * x
is the equation for a hyperbolic
curve.
( b + x )
|
 the key to
using the M&M equation is
understanding what the Km
[Michaelis Constant] is... the key to
using the M&M equation is
understanding what the Km
[Michaelis Constant] is... |
| |
|
|
Km - the Michaelis Constant
What does it tell us
|
...it's applicable to enzyme
reactions involving a single substrate
...it's "inherent
tendency" of reactants to
interact chemically for that reaction
...it's a constant that is
independent of [ES] and is defined
by [S]
...it's a mathematical
interpretation of an enzyme reaction's
kinetics
...it's a measure of how efficiently an
enzyme converts a substrate to product
...it's the substrate
concentration... when
enzyme velocity is equal to
½
Vmax
|
|
thus, when
V
=
½ Vmax
v = Vmax
[S]
thus
Vmax
= Vmax [S]
(Km)
+ [S]
2
(Km)
+ [S]
|
|
Solve
1
= [S]
&
rearrange
2
(K m) +
[S]
Km
+ [S]
= 2 [S]
thus Km =
[S]
|
native values
for Km's 10-1
to 10-7
M
"average"
Km is 10-4
M |

|
Km
is a
characteristic physical
property for
each and every different enzyme.
it
is independent of [E] and is
independent of [S]
it measures "relative
affinity" of an enzyme for its
substrate...
|
|
suppose there's more
than 1 possible substrate for a
particular enzyme, say...
kinases
- enzymes that transfers phosphate
groups from high-energy donor
molecules, such as ATP, to specific
substrates, each enzyme having its
own Km...
ex: one enzyme
with 2 substrates each with
following Km's = 1 mg &
25 mg
thus, one takes less substrate to
reach same rate…
½
Vmax rate
figure*
many enzymes have individual steps
in a complex reaction
sequences,
each step has its own Km's…
i.e., Km is a complex
function of many individual rate
constants
not all enzymes
are treatable by M & M kinetics…
most regulatory enzymes (multi-subunits)
are not treatable by M&M
kinetics.
|
Km is the
concentration of substrate which
permits the enzyme to
achieve half Vmax.
An enzyme with a high
Km has a low affinity for
its substrate, and requires a greater
concentration of substrate to achieve Vmax.* |
 |
|
|
Some ways to determine
Km... the [S] at ½ Vmax
|
|
1. by
extrapolation from a graph* of a M
& M standard
curve
v vs. [S]
|
|
|
 |
 |
|

Practical
uses of Km:
determining effect on other molecules on
an Enzymes' Km (shape):
Enzyme
Inhibition... reducing
reaction rates via binding of non-substrate
molecule
2 classes of enzyme
inhibitor molecules:
1. IRREVERSIBLE - inhibitor molecule
can not be easily removed from
enzyme,
thereby
reducing the total number
of working enzyme molecules (lowers Vmax).
i.e, enzyme is physically altered by binding
of inhibitor - reducing its amount.
ex: alkylating agents like iodoacetamide (bind to CYS-SH’s)
organophosphorous compounds- nerve gases (bind to SERs)
some
antibiotic drugs, such as penicillin, form covalent links to
enzyme active site.
2. REVERSIBLE - enzyme activity may be
restored by overcoming the effect of the inhibitors
and are thus treatable by M & M kinetics
2 major types of reversible
inhibitions...
a.
COMPETITIVE
b. NON-COMPETITIVE

First
let's look at Reversible Enzyme Inhibition as it
is treatable by M&M Kinetics:
REVERSIBLE
COMPETITIVE INHIBITION...
inhibitor binds to E
& forms an [EI] complex* at the active site
inhibitor often looks like substrate... fools active site &
binds.
extent of inhibition is concentration dependent, [inhibitor is often at fixed
conc]
thus it can be overcome* if
[S] is very high,
i.e., [S] >>> [I]
one classical example is
malonic
acid
inhibition
of SDH*
easy to demonstrate is via graphical plots*
►
shows Vmax is SAME,
but Km
value is increased

REVERSIBLE
NON-COMPETITIVE
INHIBITION...
inhibitor binds to E,
forms an [EI] complex* not at the active site
inhibitor often bears
no structural relationship
to substrate
removes a net amount of active enzyme, i.e.,
lowers total [E]
i.e., it
can NOT be
overcome, even if
[S] is very high
easy to demonstrate via graphical
plots*
►
shows Km
is SAME
& Vmax
is different figure*
One can also measure binding kinetics in
facilitated diffusion and
signal molecules using Michaelis-Menten
analyses. figure*
 <--
examples of reversible competitive
inhobition <--
examples of reversible competitive
inhobition
Examples
of Competitive Enzyme
Inhibition and
some Mechanisms of Drug Action
ACE Inhibitors
- drugs that bind to
the ACE enzyme active site
& reduces its activity.
ACE
- Angiotensin
Converting Enzyme: a
proteolytic enzyme that cuts
Angiotensin
I,
a polypeptide of 10 amino
acids, into Angiotensin
II (of 8 amino
acids).
Angiotensin*
II
promotes hypertension
( high
blood pressure - HBP
) via vasoconstriction
in 1960's John Vane discovered TEPROTIDE in Brazilian
pit viper venom, a
nonopeptide (of 9aa = Tyr-Trp-Pro-Arg-Pro-Gln-Ile-Pro-Pro)
which can functions
as a competitive
inhibitor* by
binding to the active site of
the ACE
enzyme.
today there are a number of
synthetic peptide ACE
inhibitors, all called "prils"...
(lisinopril,
captopril,
trandolapril,
moexipril,
ramipril, etc...
another competitive
inhibitor example...
Viagra*
 <-- Irreversible
enzyme inhibition
<-- Irreversible
enzyme inhibition
2.
IRREVERSIBLE ENZYME
INHIBITION... Mechanism of Action of some
Inhibitors...
a.
Sarin gas*: a nerve gas agent
forms a covalent link to serine at active site of
enzymes
b. Antibiotics
- a natural molecule (often made by bacterial
cells) that can kill other
bacterial cells (& without hurting eukaryotic
cells, which are insensitive)
ex:
Penicillin
- any one of a group of
antibiotics derived from the fungus
Penicillium.
The action
of natural penicillin was
accidentally discovered (1928) by Scottish
bacteriologist
Alexander Fleming
(1881-1955).
Howard Florey
(1898-1968) & others noted anti-bacterial
effect.
Penicillin
is an
analog-like molecule structurally similar to bacterial
peptidoglycans, which
irreversibly binds at active
site of peptidoglycan
transpetidase [cross-linking*]
thereby reducing
the enzyme's activity, weakening bacterial
cell walls that
results in rupturing & cell death.
 < --link to enzyme
nomenclature
< --link to enzyme
nomenclature
CLASSIFICATION OF ENZYMES
Enzyme Commission -
IUBMB International Union Biochemistry
& Molecular Biology
some
history of enzyme nomenclature by the
IUBMB
4 digit Numbering
System [1.2.3.4.]
established by Enzyme Commission 1958
1st
#... one of 7 Major
Classes of Enzyme Activity* [EC 1.11.1.6]
2nd
#... a subclass (e.g.,
type of bond acted upon)
3rd
#... a subclass (e.g.,
group acted upon, cofactor required, etc...)
4th
#... a serial
number… (e.g., order in which enzyme was added to
list)

 next
lecture - Metabolic Design next
lecture - Metabolic Design
SKIP ALL THE MATERIAL BELOW
Some specific Examples of
Native Enzyme Inhibition:
1.
Irreversible Enzyme
Inhibition & Mechanism of Action of some
Inhibitors...
a.
Sarin gas*: a nerve gas agent
forms a covalent link to serine at active site of
enzymes
b. Antibiotics
- a natural molecule (often made by bacterial
cells) that can kill other
bacterial cells (& without hurting eukaryotic
cells: they're insensitive)
ex:
Penicillin
- any one of a group of
antibiotics derived from the fungus
Penicillium.
The action
of natural penicillin was
accidentally discovered (1928) by Scottish
bacteriologist
Alexander Fleming
(1881-1955).
Howard Florey
(1898-1968) & others noted anti-bacterial
effect.
Penicillin
is an
analog-like molecule structurally similar to bacterial
peptidoglycans, which
irreversibly binds at active
site of peptidoglycan
transpetidase [cross-linking*]
thereby reducing
the enzyme's activity, weakening bacterial
cell walls that
results in rupturing & cell death.

2a.
Competitive
Enzyme Inhibition and some
Mechanisms of Drug Action
ACE Inhibitors - drugs that bind to the enzyme's active
site & reduces its activity.
ACE - Angiotensin Converting Enzyme: a proteolytic enzyme that
cuts Angiotensin
I,
a polypeptide of 10 amino acids into Angiotensin II (of 8
amino acids). figure*
Angiotensin
II promotes
hypertension ( high blood
pressure - HBP ) via vasoconstriction
in 1960's John Vane discovered
TEPROTIDE in Brazilian pit viper venoms,
a
nonapeptide (9aa = Tyr-Trp-Pro-Arg-Pro-Gln-Ile-Pro-Pro) which can
functions as
a competitive
inhibitor*
by binding to the active site of the ACE enzyme.
today there are a number of synthetic peptide ACE
inhibitors, all called "prils"...
(lisinopril,
captopril,
trandolapril,
moexipril,
ramipril, etc...
another competitive inhibitor example...
Viagra*
 <-- link to enzyme nomenclature
<-- link to enzyme nomenclature
SKIP
all of the material below - move to this link
Kinetic
Mechanisms of REGULATION of Protein &
Enzyme Rates…
Some
approaches commonly employed by cells...
1. by controlling
number of enzyme molecules present (gene
action)
2. by sequestering (compartmentalizing) – for example into lysosomes, mitochondria
3.
by
converting inactive peptides to active enzymes
- often involves hormones, acid hydrolysis
and/or digestive proteases
- pancreas makes zymogens...
(an inactive enzyme large precursor)
ex: pepsinogen* &
trypsionogen &
chymotrypsinogen
enterokinase*, an aminopeptidase from the
lining of small intestine...
it hydrolyzes trypsinogen to trypsin
(active form), which itself
hydrolyzes chymotrypsinogen to chymotrypsin.

4.
regulation by adjusting
reaction rates of
existing enzyme (often via... M&M
kinetics)
a) STOICHIOMETRIC controls - limit amount a reactant (substrate) present
b)
ALLOSTERY -
[allosteric kinetics...
akin to noncompetitive inhibition kinetics]
- binding of a regulator ligand results in a
change of 3o/4o conformations
- common in multimeric proteins and
enzyme complexes
- allosteric proteins have 2 binding sites:
active site = substrate
allosteric site = regulator ligand
1)
aspartate transcarbamylase initial
enzyme in pyrimidine synthesis-
ecb 4.40*
binding of CTP favored tight
conform = inactive state = feedback inhibition
- active form = +
catalysis
&
inactive conformation =
-
catalysis
- ligand often
serves as substrate, activator, or inhibitor
(or all three)

Some additional examples
of
Ligand induced Allostery...
2) Cooperative Binding: binding of one ligand affects
subsequent bindings
if + = enhances subsequent
bindings
if - = sequential bindings
are inhibited
ex:
HEMOGLOBIN: binding of 1 O2 to
a heme = Δ in local conformation
enhances
"Km" of binding
of additional O2 to other
subunits chains (mcb3.30) & cooperative binding (McKee 5.41*)
3) Ligand-induced
binding activations of catalysis:
a. inactive PKA is
activated by cAMP...
binding of cAMP
induces Δ in conformation, so that a tetramer dissociates
into 2 active monomers & a dimeric regulatory
subunit (ecb16.25*)
thus a hormone signals --> cAMP --> active
PKA dimer
without cAMP we have an inactive tetramer
b. GroEL chaperonin:
is made of 2 multi-subunit
rings
(ecb 4.10*)
binding of ATP
& GroES to GroEL results in a tight peptide binding
complex,
which closes the folding cavity allowing
efficient folding of nascent proteins

3. Calmodulin*: a Ca binding messenger protein:
functions as messenger protein altering targets
Calmodulin,
is a helix-loop-helix protein, has 4 Ca
binding
sites...
[MOVIE*]
4 Ca ions bind = Δ in
conformation - now binds target proteins
= altering its activity>
Calmodulin modulates processes as inflammation,
apoptosis, smooth muscle contraction, etc...
4. GTPase super family:
a group of hydrolase
enzymes that bind & split GTP
switching between
active/inactive forms via signal transduction.
(includes Ras
& G-proteins)
GTP Binding Proteins (G Proteins)
are Active when GTP is
bound to protein
ecb
15.15 & 15.16*
Inactive when GTP is hydrolyzed to
GDP
serve as
molecular switches, esp. cell signaling.
5.
COVALENT MODIFICATION of existing enzymes often
involves...
a.
addition of P to an inactive enzyme --> activate enzyme via P transfer
[reversible phosphorylation changes
protein conformation]
b. done
by - PROTEIN
KINASES, which transfer
P from ATP
ecb 15.15*
tyrosine kinases add P
to TYR residues of
enzymes de-activating them
serine/theronine kinases add P to SER or THR residues
- PROTEIN PHOSPHATASES... dephosphorylate, thus
inactivating

Net RESULTS of Protein Regulatory
mechanisms...
all
Help Control & Regulate Metabolism.
feedback inhibition (negative allosteric
regulation)
an initial enzyme
is inhibited by end product
prevalent in the amino acid biosynthetic
pathways -
figure*
► Primary mechanism of action is altering enzyme's
conformation
either negatively or
positively*
SKIP EVERYTHING BELOW THIS POINT
|
SKIP ALL THE MATERIAL BELOW
Some specific Examples of
Native Enzyme Inhibition:
1. Irreversible Enzyme
Inhibition & Mechanism of Action
of some Inhibitors...
a.
Sarin gas*: a nerve gas
agent forms a
covalent link to serine at active
site of enzymes
b. Antibiotics
- a natural molecule (often made by
bacterial cells) that can kill other
bacterial cells (& without hurting
eukaryotic cells: they're insensitive)
ex:
Penicillin
- any
one of a group of antibiotics derived from the
fungus Penicillium.
The action
of natural
penicillin was accidentally
discovered (1928) by Scottish
bacteriologist
Alexander
Fleming
(1881-1955).
Howard
Florey
(1898-1968) & others noted
anti-bacterial effect.
Penicillin
is
an
analog-like molecule structurally
similar to bacterial
peptidoglycans, which
irreversibly binds at
active site of peptidoglycan
transpetidase [cross-linking*]
thereby reducing the
enzyme's activity, weakening
bacterial cell walls that
results in rupturing & cell
death.

2a. Competitive Enzyme
Inhibition and some
Mechanisms of Drug Action
ACE Inhibitors - drugs that
bind to the enzyme's active site &
reduces its activity.
ACE
- Angiotensin Converting
Enzyme: a
proteolytic enzyme that cuts
Angiotensin I,
a polypeptide of 10 amino acids into Angiotensin II
(of 8 amino acids).
figure*
Angiotensin
II promotes hypertension ( high blood pressure
- HBP ) via vasoconstriction
in 1960's John Vane discovered
TEPROTIDE in Brazilian pit viper
venoms, a
nonapeptide (9aa = Tyr-Trp-Pro-Arg-Pro-Gln-Ile-Pro-Pro) which
can functions as
a competitive
inhibitor* by binding to
the active site of the ACE enzyme.
today there are a number of synthetic
peptide ACE
inhibitors, all called "prils"...
(lisinopril,
captopril,
trandolapril,
moexipril,
ramipril, etc...
another competitive inhibitor
example...
Viagra*
 <-- link to enzyme
nomenclature
<-- link to enzyme
nomenclature
SKIP all of the material below -
move to this link -
Kinetic
Mechanisms of REGULATION of Protein
& Enzyme Rates…
Some approaches commonly
employed by cells...
1. by controlling
number of enzyme molecules
present (gene
action)
2. by sequestering (compartmentalizing) – for example into
lysosomes,
mitochondria
3.
by converting inactive
peptides to active enzymes
- often involves hormones, acid
hydrolysis and/or digestive proteases
- pancreas makes
zymogens...
(an inactive enzyme large precursor)
ex: pepsinogen* &
trypsionogen &
chymotrypsinogen
enterokinase*, an aminopeptidase from
the lining of small intestine...
it hydrolyzes trypsinogen to trypsin
(active form), which itself
hydrolyzes chymotrypsinogen to
chymotrypsin.

4.
regulation by adjusting reaction
rates of
existing enzyme (often via... M&M kinetics)
a)
STOICHIOMETRIC
controls -
limit amount a reactant (substrate) present
b)
ALLOSTERY -
[allosteric kinetics...
akin to noncompetitive inhibition
kinetics]
- binding of a regulator ligand
results in a change
of 3o/4o conformations
- common in multimeric
proteins and enzyme complexes
- allosteric proteins have 2 binding
sites: active site = substrate
allosteric
site
= regulator ligand
1)
aspartate
transcarbamylase initial enzyme in
pyrimidine synthesis-
ecb 4.40*
binding of CTP favored
tight conform = inactive state = feedback
inhibition
- active form =
+
catalysis
&
inactive conformation =
- catalysis
- ligand
often serves as substrate, activator,
or inhibitor (or all three)

Some additional
examples of Ligand induced
Allostery...
2) Cooperative
Binding: binding of one
ligand affects subsequent bindings
if + =
enhances subsequent bindings
if - =
sequential bindings are inhibited
ex:
HEMOGLOBIN: binding of 1 O2 to a heme = Δ in local conformation
enhances "Km" of binding of
additional O2 to other
subunits chains (mcb3.30) &
cooperative binding (McKee
5.41*)
3)
Ligand-induced binding
activations of catalysis:
a. inactive PKA
is activated by cAMP...
binding of cAMP
induces Δ in conformation, so that a tetramer
dissociates
into 2 active monomers & a dimeric
regulatory subunit
(ecb16.25*)
thus a hormone signals --> cAMP
--> active PKA dimer
without cAMP we have an inactive
tetramer
b. GroEL
chaperonin: is made
of 2 multi-subunit
rings
(ecb
4.10*)
binding of ATP
& GroES
to GroEL
results in a tight
peptide binding complex,
which closes the folding cavity allowing
efficient folding of nascent proteins

3. Calmodulin*: a Ca binding messenger protein:
functions as messenger protein altering
targets
Calmodulin,
is a helix-loop-helix protein, has 4
Ca binding
sites...
[MOVIE*]
4 Ca ions bind =
Δ in conformation - now binds
target proteins = altering its
activity>
Calmodulin modulates processes as
inflammation, apoptosis, smooth muscle
contraction, etc...
4. GTPase super
family: a group of hydrolase enzymes
that bind & split GTP switching
between
active/inactive forms via signal
transduction. (includes Ras & G-proteins)
GTP
Binding Proteins (G Proteins)
are Active when
GTP
is bound to protein
ecb
15.15 & 15.16*
Inactive when GTP is hydrolyzed
to GDP
serve as molecular
switches, esp. cell signaling.
5.
COVALENT
MODIFICATION of existing enzymes
often involves...
a.
addition of P to an inactive enzyme
--> activate enzyme via P
transfer
[reversible phosphorylation
changes protein conformation]
b. done
by - PROTEIN KINASES,
which transfer P from ATP
ecb 15.15*
tyrosine kinases add P to TYR residues of
enzymes de-activating them
serine/theronine kinases add P to SER or THR residues
- PROTEIN PHOSPHATASES...
dephosphorylate, thus inactivating

Net RESULTS of Protein
Regulatory mechanisms...
all
Help Control & Regulate
Metabolism.
feedback inhibition (negative
allosteric regulation)
an initial
enzyme is inhibited by end product
prevalent in the amino acid biosynthetic
pathways -
figure*
► Primary mechanism of action is
altering enzyme's conformation
either negatively
or positively*
|
SKIP
THIS
Derivation of Michaelis-Menten
Equation
k1
k3
E + S ↔
ES
↔
E + P
k2
k4
rate
limiting step is
ΔP/Δt
= k3[ES]
(&
ΔP/Δt
=
v
)
1.
Rate of formation of ES complex
ΔES /Δt
=
k1 [E T -
ES] [S]
2.
Rate of destruction ES complex
ΔES /Δt
= (k2 +
k3) [ES]
3.
Steady State Equilibrium
k1 [ET
- ES] [S]
= (k2+
k3) [ES]
4.
Michaelis Constant
(Km)
(k2 +
k3)
=
[ET
- ES]
[S]
(k1)
[ES]
down
Km
= (k2 +
k3)
=
[ET
- ES]
[S]
(k1)
[ES]
5.
Solve for [ES ] [ES]
=
[ET]
[S]
(Km) +
[S]
6.
Substitute in above
ΔP/Δt =
k3
[ES]
v = k3
[ET]
[S]
Km
+ [S]
7.
Substitute Vmax for
k3 [ET] v =
Vmax
[S]
Km +
[S]

|
|
Recall: the
definitions of enzyme activity:
a
way of standardizing the expression of the
physical properties of an enzyme... |
most often measured by relative rate
substrate ---> product
|
1
international unit of enzyme ACTIVITY is that amount enzyme
protein which converts
1 umole substrate per min at 25oC
& optimal pH
|
1 unit SPECIFIC ACTIVITY
# units per mg of protein present
(e.g.,
37umole/min/mg protein = 37 units/mg)
|
1 unit MOLECULAR ACTIVITY
# units per umole of purified
enzyme
(e.g., 12
units/umole of enzyme)
|
| |
|
|
|
|
|
h
|