Early
chemical analyses described Two Classes of
Proteins - SIMPLE & COMPLEX
SIMPLE PROTEINS: on
acid hydrolysis yields
only
alpha-L
amino acids*:
Chemical analysis of
proteins:
historically based on SOLUBILITY
of PROTEINS,
via the chemical
properties of isolated proteins...
1. Albumins - soluble in pure water (distilled);
are globular in shape; includes many
enzymes
2. Globulins
- soluble
in dilute aqueous solutions (with some ions); insoluble in pure distilled
water
3. Prolamins - insoluble in water; but soluble
in 50% to 90% simple
alcoholic solutions
4. Glutelins - insoluble in
most solvents; but soluble in dilute acids/bases
Later classification was based upon
amino acid content not upon
solubility:
5.
Protamines - small
MW proteins with 80%
Arginine & no Cysteine (these bind to DNA)
6.
Histones - have high # basic aa's - 90% Lys, Arg, & His : form nucleosomes in DNA.
7. Scleroproteins - are
insoluble in most solvents and have a fibrous
structure
- architectural proteins of cartilage &
connective tissue
Collagen - high Glycine, Proline, & no Cysteine; when boiled makes gelatin.
Keratins - proteins of skin & hair, high
basic aa's (Lys,
Arg, His),
but w some Cys
 images & examples of simple
proteins
images & examples of simple
proteins
Complex Proteins:
on hydrolysis yield amino acids +
other molecules
Lipoproteins - (+ lipids)
blood,
membrane, &
transport proteins
Glycoproteins -
(+ carbohydrates)
antibodies, cell
surface proteins
Nucleoproteins -
(+ nucleic acids)
ribosomes &
organelles |
 |
Some common protein terminology:
dipeptide
= 2 amino acids;
tripeptide = 3 amino acids
oligopeptide
= short chain of amino acids
(2-20);
peptide < 50
aa;
polypeptide =
few to many amino acids (up to
300); MW ≈ 10,000
protein = polypeptide
with well defined 3D structure

 thought
question? thought
question?
... based on 20 amino
acids what's max possible number of
different proteins*
|
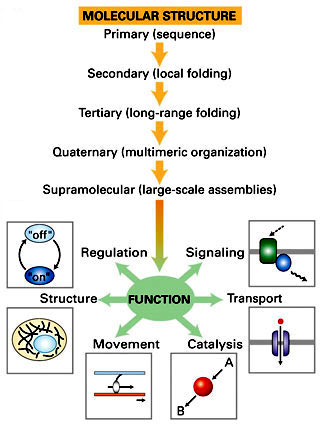
Structure of
Proteins
the Variety
of Protein Structures may be INFINITE...
- proteins are made of some 20 alpha-L amino
acids
-
average protein has
some 300-500 amino acid's &
a MW of 35kD
to 55kD
- thus a PROTEIN
of 300 aa's could have 20300 different linear arrays of amino
acids

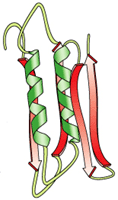
4 levels of
dynamic protein
structure* are
recognized
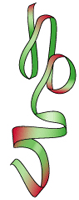 |
primary
- linear sequence
of aa's
will define properties of the protein
|
 |
secondary
- regular, recurring orientation of aa
in a peptide chain due
to H-bond |
tertiary
- complete 3-D shape of a
peptide
due to weak
electrostatic forces |
quaternary
- spatial relationships between 2 or
more different peptides or subunits |

Primary
sequence…
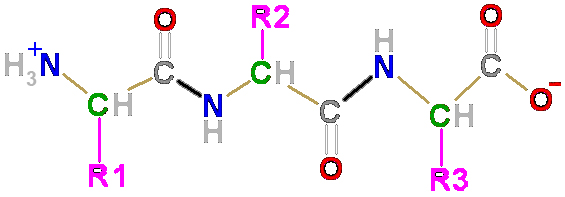
|
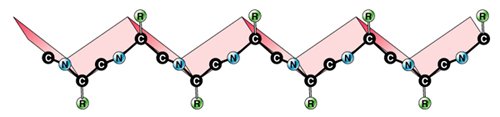
Linear sequence of
amino acids in a polypeptide
repeated peptide bonds form the back
bone of the polypeptide chain
R side
groups project outward on alternate
sides along a zig-zag backbone
|
Chain...
one end of polypeptide chain has a
free (unlinked) amine group:
N-terminus
other
end has a free (unlinked) carboxyl
group: C-terminus
N-C-C-N-C-C-N-C-C-N-C-C-N-C-C-N-C-C
|
Size…
a protein's size is specified by its MASS
(MW in Daltons = 1 amu)
average MW of
all the individaul amino acids is
≈ 113
Da
thus if a protein is determined to
have a mass of 33,900 Da
≈ 300
amino acids
average yeast protein =
52,728 Da [52.7 kDa] and thus is
about 466 amino acids
|
Protein
Primary Sequence today is often
determined by reading the GENOME Sequence???
by looking for RNA initiator
codon (AUG)
& its DNA complement (TAC) in
Human genome.
|
Protein
function is derived from the
3D structure (conformation), which is specified by
the primary amino acid
sequence and its local environs
interactions.
|
 [lysozyme*]
[lysozyme*]
|
| |
some important consequences... of
Primary Structure
amino acid Sequences…
|
Polymorphism...
proteins may vary
in their primary amino acid sequence,
have a
different structure, but still exhibit
the same
catalytic activity...
ex:
peroxidase family*... H2O2
--> 2
H2O + O2
inter-specific:
between species [each have
different aa
sequences]
intra-specific:
within a species [ liver
vs. kidney ]
ex:
lactic dehydrogenases (pyruvate
--> lactic acid) -
LDH isozymes*
|
Invariants... don't
vary significantly in aa sequence
[insulin]
ex:
ubiquitin* (proteasomes -
96% eukaryotic sequence
universality)
&
histones*
(chromosomes
- few
sequence difference
among eukaryoted
|
Site Specificity… unique
sequences determine intra-cellular
location & function
ex:
signal sequence* for
protein targeting, prosthetic binding sites, etc…
|
Families of Proteins:
different structures but with related
functions; having evolved
from
a single ancestral
protein & may have up to
30%+ commonality
of sequence...
serine
proteases (trypsin,
chymotrypsin,
elastase) all
have
SER*
at active site, resulting in
nucleophilic hydrolysis of peptide
bonds.
|
Mutation...
change in primary amino acid sequence
= defective protein -
Sickle
Cell Trait*
a non-polar amino acod replaces a
polar charged amino acid altering
shape |
| Visualization... some
common ways primary sequences are
depicted ecb 4.12*
|
 |
Secondary
structure -
is a well defined periodic structure making up
some 30% of a protein's structure
Alpha helix*
[ animation* ]
described by Linus Pauling (1954
Nobel)
using
X-ray diffraction*
| rigid rodlike cylinder
around long axis core |
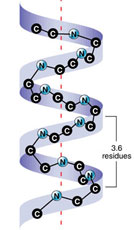 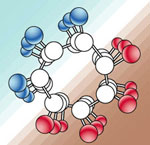
click for dimensions*
|
| R-groups radiate outward |
3.6 aa per 360o
turn
[1.5Ao/residue -
5.4Ao/turn] |
| single repeat turn of
helix (360o) = 0.54 nm |
| right handed helix forms
counterclockwise |
| helix formed from H-bond
interactions |
| H of N (of one
aa) &
-C=O
(of 4th aa away) |
| How much
of typical protein is in
alpha helix? |
|
about 30%
of a protein is in alpha helical shape. |

|
|
|
|
Secondary
structure- animation* ecb
4.13*
(fig
3.5 & McKee
5.19)
short
segments (3-10 residues) CONNECT
LATERALLY by H-bonds
= pleated
sheets,
e.g., a
linear extended ZIG-ZAG pleated
sheet
- intra- &
inter-chain
⅓ of typical
protein structure
is also in beta
sheets. |
can be parallel and antiparallel -
ecb 4.17*
resist pulling (tensile)
forces = strength of
silk fibers:
model = fibroin |
α/β
regions
combine to help establish initial
shape in proteins - ribbons
& sheets*
non- α/β regions
include hinges, turns, loops,
etc = flexibility
β-TURNS - a region of 3 or
4 amino acids that redirect
backbone;
mcb 3.6*
involves 4 residues: 1st & 2nd =
PRO in cis; 3rd = GLY;
& a 4th;
Proline
Turns - are due to
either a cis
configuration of proline ring. |
|
|
|
|
|
Polypeptide
folding MOTIFS & Domains
are conserved Super-Secondary
Structures...
Motifs are 3D combinations of 2nd
structure that appear in a variety
of
other proteins and
enzymes which can have a unique
function.
vs: a
Domain
is a folded section and has a
discete function in a
protein.
|
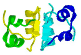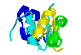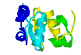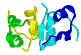
|
motifs
are recurring arrangements
of α-helix and/or
β-sheets, & αβ-motifs,
can occur in different proteins
with/without similar functions?
|
►
Examples of alpha
helix & beta sheet motifs
often within a protein homeodomain:
a
homeodomain is a conserved amino acid
structure domain that
binds to DNA & functions as a DNA
transcription factors...
EF hand...
two short α-helices connected by a
loop with a Ca+2 ion binder is a
homeodomain
of 60 amino acid helix-turn-helix
DNA-binding domain
fig 3.9b* - Animation*
Zinc
finger...
1 α and 2 β (β-α-β) strands
with antiparallel orientations form
'fingers'
bound by Zn ion
that often link to DNA (RNA)
fig 3.9c* Animation*
Coiled
coil... helicies,
where the hydrophobic
amino acids in one helix wind
together
forming a coil with others; also
called leucine
zippers* due to
high [leu]:
common to transcription factors -
coiled-coil
anim*
[ a synthetic coil ]
 alpha helix & beta sheet
motifs are often common in
transcription factors
alpha helix & beta sheet
motifs are often common in
transcription factors |
Tertiary level 
is level most responsible for 3-D orientation of
proteins in a cell's internal
environment...
it's the most thermodynamically
most stable conformation of
a protein... and is due to
–
weak
non-covalent molecular interaction
forces* [panel-2.3-
weak molecular forces]
- hydrophobic interior & hydrophilic
exterior favors globular shapes
- in
enzymes active site made via these weak
bonds (ecb
fig 4.32)
- strongest 3' force is covalent S-S
bridges... animation - ecb 4.30* - [Home
Perms]
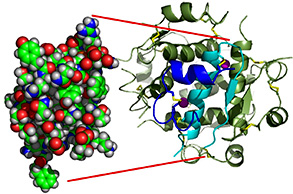
some examples of 3D structure in proteins:
Lysozyme* MW
14,600 enzyme; egg
white & human tears pdb --> lysozyme
124 aa's with 4
S-S;
that hydrolyses polysaccharies
in bacterial cell walls = bactericidal
agent
Myoglobin
MW
16,700 - animal muscle
protein - stores O2
pic
Cytochrome-C MW 12,400 - heme binding
single polypeptide
pic
of 100 aa
in ETS of mitochondria
deoxyribonuclease MW 34,000 enzyme of 262 aa
w 2 S-S
pic
ecb fig 4.11* - Scientific Animations -
Molecular-Eye-Candytake-a-look

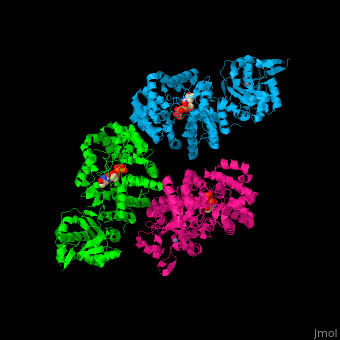
QUATERNARY Level: multiple
polypeptides each with a
3-D conformations = final shape
Some Common Quaternary
Level Protein Shapes & Assemblies...
animation*
1. dimers
- self
recognizing symmetrical regions -
bind together @ identical binding sites
[ Catabolite
Activator Protein*
] homodimers - 2 identical subunits
heterodimers -
non-identical subunits
(as in reverse
transcriptase)
2. tetramers - 4 identical subunits... ex:
neuraminidase
- ecb 4.23b* and hemoglobin*,
3. more ex: RNA polymerase and
ASP-trans-carbamoylase
Multi-Enzymes
Complexes :
pyruvate
dehydrogenase
picture* &
pic
ATP synthase
figure*

MULTIMERIC PROTEIN
COMPLEXES can have very large Quartnerary-like Structure...
and form very large MACROMOLECULAR ASSEMBLIES...
( > 1 mil Da in mass ),
30-300 nm in
size,
& 10-100 individual
peptides.
other examples include:
1. mRNA
transcription complex (some 60 proteins -
figure*)
2. Molecular Machines
of several types - see
mcb/5e - table
3.1*
We will look at some of these in greater
detail later...
Animations
of molecular motors can enhance science14
min
3. virus - infective
molecular complexes of nucleic acid &
proteins
ex: coronavirus
Protein
Data Bank (1971) is
a database repository of 3D structures
of large
biological molecules, as proteins and
nucleic acids:
- Protein
database files animation*3
min

Assessing a Protein's 3D Conformation
is crucial to knowing its Biological
Function...
DENATURATION:
in 1931 a Chinese biochemist, Hsien
Wu, showed loss
of protein shape
(via protein
denaturation) resulted in loss of protein
function...
A loss of 3D conformation is often
caused by Δheat, ΔpH,
+organic
solvents, +detergents.
Anything that disturbs 20/30/40 level forces can result
in denaturation.
How does one monitor protein
denaturation?
Conformational changes in proteins
invariably
affect
several of its chemical and physical
properties, such as ultraviolet (UV)
absorbance,
fluorescence, viscosity, sedimentation
coefficient, optical rotation,
reactivity of sulfhydryl
group
bindings, and enzyme activity.
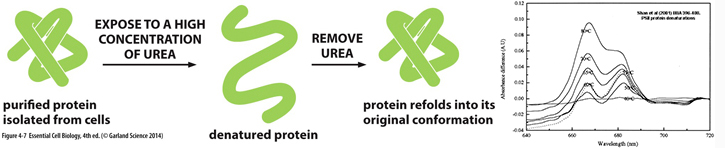
Common protein conformations
& shapes...
|
NATIVE Protein
3D-CONFORMATION is the…
3-D SPATIAL
ORIENTATION that is
MOST thermodynamically
STABLE
and has the lowest free energy
expenditure (likely forms spontaneously)
the 3 most COMMON
PROTEIN 3D-CONFORMATIONS
include...
HELIX
- a spiral staircase-like shape
FIBER - elongated bound
monomers
GLOBULAR - roughly a
sphere
|
the Native Conformation of
MOST ENZYMES & soluble
proteins is GLOBULAR:
an interior pocket of hydrophobic amino
acids
an exterior surface of hydrophilic amino
acids
- maximizes
the number H-bonds that form
ecb 4.5*
|
|
non-covalent
bonds, H-bonds, hydrophobic & hydrophilic
interactions,
&
strong covalent bonds (as peptide
bonds & disulfide
bonds)
results in a
great variety
of protein shapes & sizes - ecb 4.10 pg
125
|
|
|
PROTEIN SHAPE... to FUNCTION PROPERLY
a protein must be FOLDED
into a unique shape,
so that it can interact with other
molecules... always
in constant motion*
Structured proteins...
such as ENZYMES,
often have either a preformed
Lock
& Key*
shape or an
Induced Fit* shape to recognize
specific ligands.
in 1894 Emil Fischer
(U. Berlin)
1st proposed enzymes bind to specific
shaped
molecules via a preformed-shaped site
(original lock-key active site hypothesis)...
However, recent evidence
indicates many proteins can functions using
unstructured regions*...
It is estimated that about ⅓ of human proteins lack a rigid,
predetermined structure,
i.e, they are INTRINSICALLY
DISORDERED (not
precisely structured).
to date about 600 unstructured regions
are identified, each with a high
hydrophilic aa
content
compared to
rigidly folded proteins. DNA sequence searches
for high hydrophilic aa regions
suggests as
much as 35% of all human
proteins may
have unstructured regions.
Some unstructured (intrinsically
disordered ) proteins with unique functions
include:
►
unstructured regions of proteins - kinesin, p53
regions, nuclear pore complex*
 Structured
shapes (Lock & Key ) favor
high specificity (smells*) & thus favor enzyme
activity.
Structured
shapes (Lock & Key ) favor
high specificity (smells*) & thus favor enzyme
activity.
Disordered
(unstructured) proteins might be best
for signaling, regulation, control functions.
How does 3D protein folding come about?
"FOLDING determines FUNCTION"
► a native folded conformation is the most stable, i.e., has a low entropy state, and is often
formed by R-group chemical
properties (size, hydrophobicity,
hydrophilicity, & ionic strength).
Folding
involves: chemical
changes leading to a native 3D
conformation:
figure*
- occurs via
orderly steps in a sequential way, each
step facilitating the next,
-
first 20 structure forms
(α &
β), then
structural motifs & assembly of
complex domains,
- followed by 30 level forces
&/or 40 shapes,
- protein folding is based upon the
chemistry of the amino acids,
6 u-sec of protein folding* + Villin folding
via 'Folding@home'
computer simulation model*
AlphaFold* has solved the
structure of a number of
proteins
Examples of folding software: 1) understanding
misfolding can be medically
important
2) protein
Synthesis & folding 3)
Covid spike
protein
4) Make
any protein shape
Native cellular
protein folding can interact with all the
other molecules in a cell and this
must
occur in a protected Folding
Environment, which
involves 2 sets of proteins that facilitate
folding:
Molecular Chaperones
- bind
and stabilize newly made unfolded
proteins preventing these
proteins from self aggregating and/or being
denatured before folding.
Chaperonins - makeup a small folding chamber into
which unfolded proteins are moved
 to provide a proper environment favoring
native folding of a protein.
to provide a proper environment favoring
native folding of a protein.
MOLECULAR
CHAPERONES - are families
of proteins that help "properly
fold" a new protein...
several chaperones
can bind to newly made proteins and include
proteins as:
Hsp70 (of cytosol
& mitoplasm), BiP (of the
E.R.), & DnaK (of bacteria).
Chaperones were 1st discovered via heat shock treatment
[temp elevations of 25o --> 32oC]
by Ferruccio Ritossa
(1962 - Italy) who heat shocked fruit
flies giving novel Chromosome puffs*
by 1980s... shown that all cells
(bacteria to humans) produce heat shock proteins
(HSPs);
► but mutant
bacteria didn't
make Hsp's and didn't
fold their proteins normally.
HSP's
are ubiquitous
to all cells - produced in response to stress (heat,
infection, etc...)
and they can act as "Chaperones" for
folding other proteins by:
1.
inhibiting
undesirable interactions with other
proteins
2.
while promoting
desirable interactions within a
folding protein...
help form stable attractions between protein
regions, while
establishing proper
conformations & preventing aggregations.
 A mis-folded
protein- in the E.R. binds to receptor proteins in
the ER lumen and
may
A mis-folded
protein- in the E.R. binds to receptor proteins in
the ER lumen and
may
initiate transcription of chaperones
gene & chaperones move to ER
ecb 15.25*
Classes
of Heat Shock (Chaperone)
Proteins & How they Work:
Heat shock proteins are
a family of proteins, produced on exposure to
stress that can perform
chaperone-folding functions by
stabilizing new proteins to ensure correct
folding or help refold
proteins damaged by stress. Many
are made constitutively throughout the life of
cells.
Hsp -40, -60, -70, -90 & -100.
Hsp
were originally named according to their
molecular weights
(Hsp-70 = 70 kilodaltons)
Hsp-40 facilitates hydrolysis of
ATP bound to another Hsp-70
Hsp-70 and associated factors grabs
proteins by an open cleft when ATP is bound to Hsp-70;
OPEN conformation has
hydrophobic pocket for an
unfolded protein...
in its ADP
conform, it closes around
protein and aids native folding... fig Hsp70*
HSP
role and overexpression in
stressed (cancer tumor) cells.*view@home
Hsp-90 receives
partially folded proteins from Hsp-70's and
other chaperones...
helps join polypeptides
into larger quaternary proteins forming
multi-subunit
proteins, such as cellular
receptors. mechanism of action of HSP-90*

HSP-60 also known as
CHAPERONIN or 'foldase' -
is a small folding CHAMBER of HSP's into
which unfolded proteins are
moved to provide a proper environment
favoring native folding...
ecb 4.10*
and
HSP-60 animation*
CHAPERONIN, a Molecular Machine well studies in E. coli...
chaperonin
proteins form a barrel
shaped structure:
made of 14 polypeptides (via GroEL gene) in
2 donut rings and a cap (via
GroES gene)
that opens an inner chamber, where a cell's
new protein enters & is folded.
barrel chamber has 2
conformations:
tight &
relaxed.
new peptides are inserted into cavity of a GroEL chamber &
conformational changes favor native
protein folding &
ATP hydrolysis = relaxed state & releases
native 3D-protein mcb7e-fig
3.17*
proteins
fold to their lowest free energy state with
amino acids aligned at equilibrium,
with thousands of options, making computer
analysis ideal to study multiple folding
options.
optional
reviews:
Molecular
chaperone-mediated protein folding
animation
Discovery of Chaperonin protein
folding by Arthur Horwich [A.L. Horwich: PNAS 96:11-37, 1999]
Online gaming
helps solve protein folding
structures

|
|
|
|
| |
| Consequences
of Misfolded Proteins their Diseases |
some diseases
can caused by mis-folded proteins:
- cystic fibrosis is
due to a mis-folded
Cl-channel protein (CF
transmembrane conductance regulator)
that interferes with flow of salts &
fluids resulting in a sticky mucus
imparing breathing. t
|
- Prion Based
Mis-folded protein diseases:
PRION: a
defective mis-folded protein agent (PrPsc) due to misfolding of
a native protein (PrPc)...
the native prion protein
is
PrPc resides on nerve cell surfaces...
defective protein
PrPsc
accumulates
forming aggregates
that lead to CJD & SE diseases.
CJD:
Creutzfeld-Jacob disease - fatal neurological disease due
to presence of the misfolded PrPsc protein.
may be acquired (...say by eating "mad
cow" tissue) or genetic
(7.5% cases).
SE:
Spongioform
Encephalopathy - vacuolation
(holes) in brain nerve tissues (often
acquired)
|
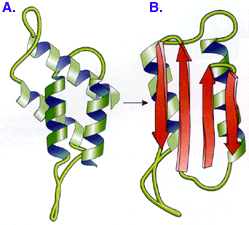
PrPc
(PrPsc) |
McGraw-Hill
Online Learning Prions*view@home
Prion -
Proteinaceous Infective Particle
Both PRION proteins can have
identical aa
sequences, but fold
differently... [are conformers* =
proteins differ in conformation]
A. normal PrPc protein... mostly
α-helix foldings -
remains soluble
B. abnormal
PrPsc protein... has 45%
β-sheet - insoluble
& is
protease insensitive forms cell
surface aggregates kills cells
in domino effect
misfolded form induces misfoldings
in normal form
|
 an
HSP-100 can
unfold proteins: How to
unfold aggregated proteins with
HSP-100 Disaggregate
an
HSP-100 can
unfold proteins: How to
unfold aggregated proteins with
HSP-100 Disaggregate |
PROTEIN DEGRADATION
(Digestion/Turnover)...& getting rid of old or
misfolded proteins
cells often contain
specialized mechanisms or pathways to digest
cell proteins...
1. to rapidly turnover proteins with short half-lives
2. to recognize & eliminate damaged or misfolded proteins
that can lead to diseases
as Alzeheimer's, and Creutzfeldt-Jacob
disease, Huntington's.
Protein
Degradation Processes:
1.
many proteins are degraded by cytosolic PROTEASES* that cut (hydrolyze) peptide bonds
2. some proteins are
degraded within LYSOSOMES via endosomes & phagocytosis fusion.
3. many proteins are
degraded by large sophisticated complexes
of proteolytic enzymes
known as PROTEASOMES in process known as
Ubiquitin-mediated
Proteolysis (UMP),
short half-life proteins
hold a signal sequence targeting proteins
for UMP
and misfolded proteins seem to be recognized
for degradation by the UMP.
may
be universal -
proteasomes occur in all eukaryotes & archaea, & in
some bacteria.
 What do proteasomes do?
What do proteasomes do?
First described by
Alfred
Goldberg &
Martin
Rechsteiner in
1980's
|
|
PROTEASOMES... a large MOLECULAR
MACHINE (ecb4e fig
7.40*)
Proteasomes
are a cell's protein
recyclers...
- they chop-up
damaged or obsolete proteins,
-
into smaller pieces - 2 to
25 amino acids,
-
which are the completely
digested into amino acids
by peptidases
|
Each proteasome
is a barrel shaped complex (2,400kD) made of
3 parts...
1) a Regulatory
Cap
of 16-18 proteins (6 with ATPase
activity)
this 19S cap only lets in
ubiquitinized proteins [purple] ,
2)
a Barrel core of 4 stacked protein
rings w protease activities
[yellow-red],
3) a
Base
cap [blue] |
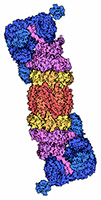
|
|
Protein
Digestion... begins when cells add a
small polypeptide (ubiquitin)
to proteins to be
degraded.
ubiquitin - globular
protein of 76 aa
(96% aa sequence homology between yeast
& human ubiquitin)
3 ubiquitin ligase enzymes [
E1, E2, E3 ]
add Ubiquitin
to proteins to be degraded;
a ubiquitinized protein is
targeted for entry into a Proteasome's*
central interior chamber,
where
proteases with
chymotrypic, tryptic,
& caspase-like
proteolytic
activity cleave the protein
into peptides. A proteasome
animation
with
the
ubiquitin being
recycled. [2004 Chemistry Nobel]
optional resources on
proteins:
Table of
the History of Protein Structure
[Table 4.2
ECB5e]
animation
review life cycle of a protein &
proteolysisflash
Games
of Science
-
non-scientist/citizen computing
projects
 next
lecture - Enzymes.
next
lecture - Enzymes.
Summary
of Chaperone folding & protein degradation
via proteosomes
SKIP
THIS PAGE & everything below
Protein
Engineering...
producing novel proteins,
with unique shapes, via artificial means...
using PROTEOMICS... to
make artificial proteins of desired sequence..
vaccine proteins
- which can bind to viral surface and
inactivate it
simplistic idea - but it's hard to make
connection from 1o to 3o
structure
1. modify existing proteins via site
directed mutagenesis*...
isolate a gene, alter
its sequence in precise way, clone
the protein product...
- can be used to study effect of one amino
acid change on 3D-folding
- often done with clinically useful proteins
to enhance efficiency (Km)
2. structure
based drug design...
make drug molecules with high binding affinity
to known proteins
[to
remove it]
use computers to design 'virtual'
drug*
to fit into a protein rendering it inactive
3. bionanotechnology...
the idea is to
exploit molecule's assembly skills to build
new nanodevices. Instead of
domesticating plants and animals, it's time to
domesticate molecules. Biology may be
able to design nanodevices that build
themselves.
►
a complex motif
example: αβ-barrel* motif
of methylmalonyl CoA mutase
DOMAINS...
a segment
of a polypeptide's 3D structure
with a characteristic shape that can evolve,
function, & exist independently of rest of
protein, that perform
specific functions...
The difference
between a Domain and a Motif is that Domains
are independently stable, while
motifs are not. A
motif can and is often part of a
Domain.
- is a
distinct modular unit or
structural elements often of 20/30 level protein structure...
- has regions that are self-forming, can fold independently,
& are self-stabilizing
- modular areas in a polypeptide of some 40 - 350 amino acids
(avg. = 100),
- may exist in multiple proteins
& may consist of several domains...
- molecular evolution uses domains as building
blocks to create new proteins w different
functions
Examples of Domains:
CAP
protein* - Catabolite
Activator Protein is a transcriptional
activator that exists as a homodimer
with each subunit comprising a ligand-binding
domain at the N-terminus, which helps
dimerization* of
the protein, and a DNA-binding domain at the
C-terminus.
Pyruvate
kinase* a glycolytic enzyme convert PEP
to PYR & makes ATP has 3
domains:
a regulatory beta, an alpha/beta binder,
& a nucleotide binding site domain.

PROTEIN FAMILIES -
Proteins
with common
evolutionary
ancestry are known as homologs often belong to a "family";
many have
identical or chemically
similar amino acids in identical sequence positions;
>30% aa sequence homology;
each may contain domains that resemble those of other
proteins.
ex:
serine proteases
ecb fig 4.21* -
proteolytic enzymes of both prokaryotes &
eukaryotes
with nearly identical amino acid sequences all with HIS-57, ASP-102,
a SER-195 at the active site
for peptide bond hydrolysis.
[ex: plasmic,
tyrpsin, elastase, tryptase, chymase, catepsin
G]
protein family relationships are often best
displayed by taxonomic cladistics
...
which is a
family tree of
shared-derived characteristics traced to
common ancestor.
ex: globins -
genes slowly diverged into animal and
plantlineages ecb
9.9*
myoglobin - monomeric oxygen binder of
muscle
hemoglobin - tetrameric oxygen binder of
blood
globin
gene-A phylogram
Today, computer modeling is used to predict function
of yet unisolated proteins
 by comparison to known sequence homologies.
[PSI*]
by comparison to known sequence homologies.
[PSI*]
Databases of
protein motifs
Other proteins
as (Aβ-protein) may misfold,
aggregate, & initiate chain reactions
that underlie Alzheimer's.
How to unfold
aggregated proteins with HSP 100
disaggregate
ex: single amino acid variants &
atherosclerosis
& ApoE
Games of Science -
non-scientist/citizen computing projects
(3D-image)
3)
model
of ACE2 fold & unfold